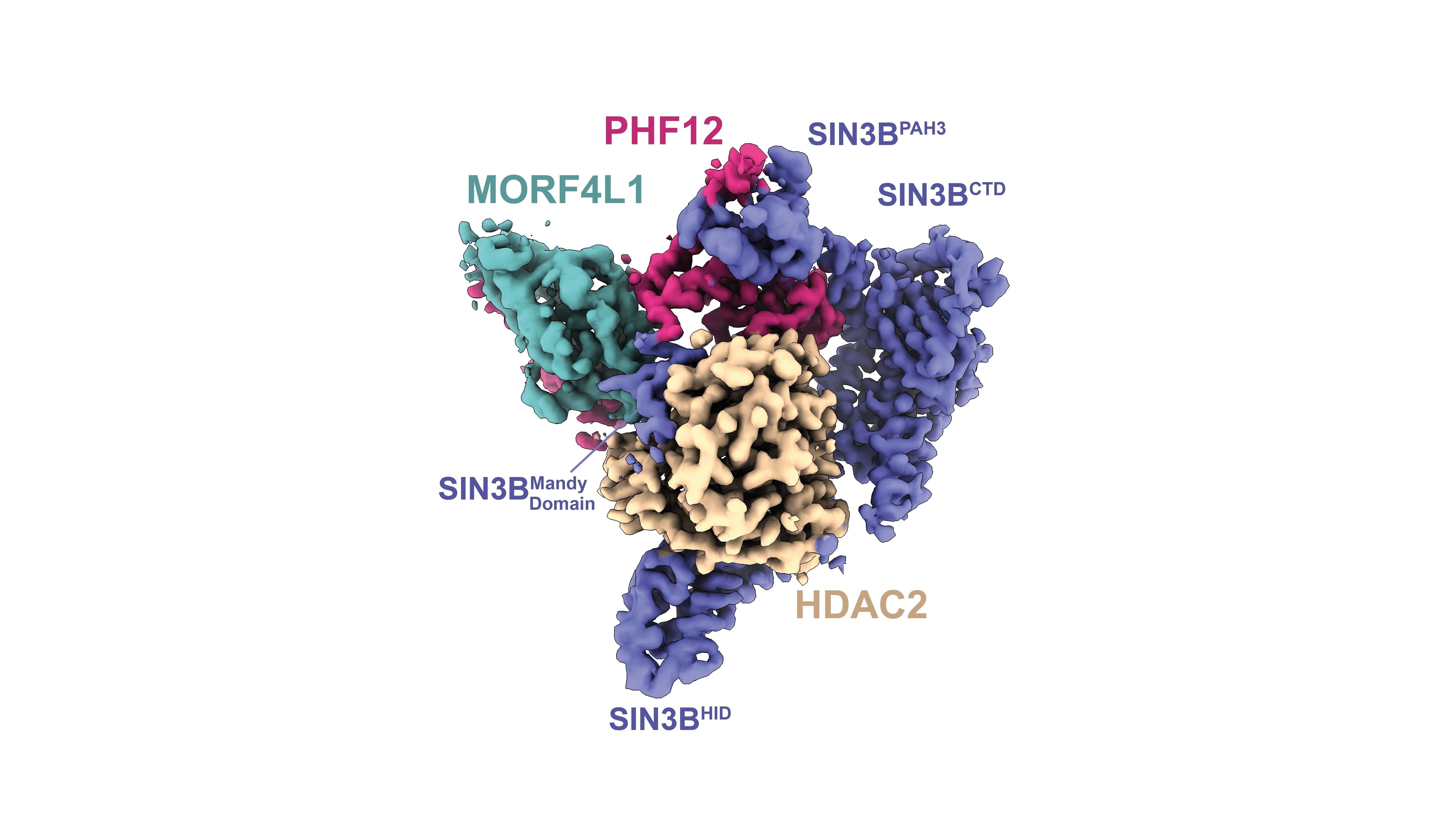
Image: High-resolution structure of the human protein complex SIN3B. Credit: Wan, Muhammad et al 2023.
Researchers have revealed, at high-resolution, the structure of a human protein complex named SIN3B, which is a ‘nanomachine’ involved in regulating cell division. Cell division is a fundamental process for life which, if it becomes uncontrolled, can lead to cancer.
The team also discovered several interaction sites within these proteins, which can be mutated in people with cancer.
This is the first time the high-resolution structure of a human protein complex of this kind has been determined.
Although the research is early-stage discovery science, the scientists at The Institute of Cancer Research, London, emphasise that their findings will have “therapeutic potential”. The high-resolution structure can now be used to design drugs inhibiting SIN3B in cancer cells that rely on this complex to evade anti-cancer therapy.
This nanomachine is involved in regulating cell division by helping to make the DNA more compact so that cell division genes get switched off, therefore restricting the rapid growth of cells.
DNA during cell division
There is so much genetic material making up the chromosomes that the DNA molecule is 2 metres in length – but it must be folded up tightly to fit into the nucleus of the cell which is about the width of spider’s silk at only 6 micrometres. The DNA is packaged up inside each cell in a structure known as chromatin, where it is wrapped around proteins called histones forming nucleosomes, which are like beads on a string.
The DNA in each cell must also be unpackaged so it can be copied and transcribed during cell division, which is a tightly regulated process. In fact, transcribing genes to make the nanomachinery required for cell division itself, is essential for the execution of this process.
The histones have linear structures coming off them – these histone tails can undergo a process known as lysine acetylation, performed by histone acetyl transferase enzymes (HATs), which helps ensure the chromatin is ready for DNA replication, DNA repair and gene transcription. Other enzymes called histone deacetylases (HDACs) counteract the role of HATs – making the chromatin more compact thereby shutting down gene transcription.
Cutting-edge microscopy
Using cutting-edge experimental techniques, such as cryo-electron microscopy and structural proteomics, Dr Claudio Alfieri’s group, in collaboration with Professor Jyoti Choudhary’s group, at the ICR were able to determine the structure of SIN3B, which surrounds the HDAC enzyme – activating it and allowing it to recognise and deacetylate nucleosomes.
The findings, published in the journal Nature Communications, show that SIN3B helps to recruit other proteins, called PHF12 and MORF4L1, which allow the nanomachine to bind to the histone tail so it can inhibit transcription.
Beyond discovering how the nanomachine assembles and does its job in regulating cell division, the researchers also showed where mutations in the proteins can occur in cancer patients – these faults stop healthy processes that suppress uncontrolled cell division and cancer.
The study, which was largely funded by a Sir Henry Dale Fellowship from Wellcome and the Royal Society, and by the ICR, which is both a research institute and a charity, will now lead to research into what happens within the cell when the researchers interfere with the normal function of the SIN3B nanomachine and see the effect on cells of removing this function. They will also look to team up with the world-leading drug discovery scientists at the ICR to see if their discoveries can be harnessed and lead to new cancer drugs.
Studying the most beautiful process in biology
Dr Claudio Alfieri, Leader of the Molecular Mechanisms of Cell Cycle Regulation Group at the ICR, said:
“The cell cycle, where cells divide and new ones are made to help our bodies repair or grow, is to me the most beautiful process in biology. But, when it becomes uncontrolled, it can be incredibly dangerous – leading to cancer. That’s why it is so important to study the biology of the cell cycle at meticulous detail and investigate everything that regulates the process.
“We’ve known about HDAC enzymes for 50 years, but we didn’t know how they specifically target the histones involved in wrapping up the DNA in our cells into a compact package in the nucleus. It’s hugely exciting to be able to see something for the first time that no one else has ever seen before.
“This work is a great achievement for us, for the two first authors of the article, Mandy Wan and Dr Reyhan Muhammad. It is also a great collaboration between both the Division of Structural Biology and the Functional Proteomics Group at the ICR.
“We now hope to better understand the role of this cell division nanomachine and see how the structure we’ve revealed can be useful to our drug discovery colleagues, who are working to find new treatments for people with cancer.”

 .
.