Mechanisms and Regulation of pre-mRNA Splicing
Professor Vlad Pena's group focuses on the structural basis of human pre-mRNA processing under normal, pathological and drug-induced conditions.
Research, projects and publications in this group
Our group focuses on the structural basis of human pre-mRNA processing, including: splicing modulation by compounds with antitumour properties, ubiquitination in splicing and DNA repair and helicases.
Catalytic activation of human spliceosomes by RNA helicases
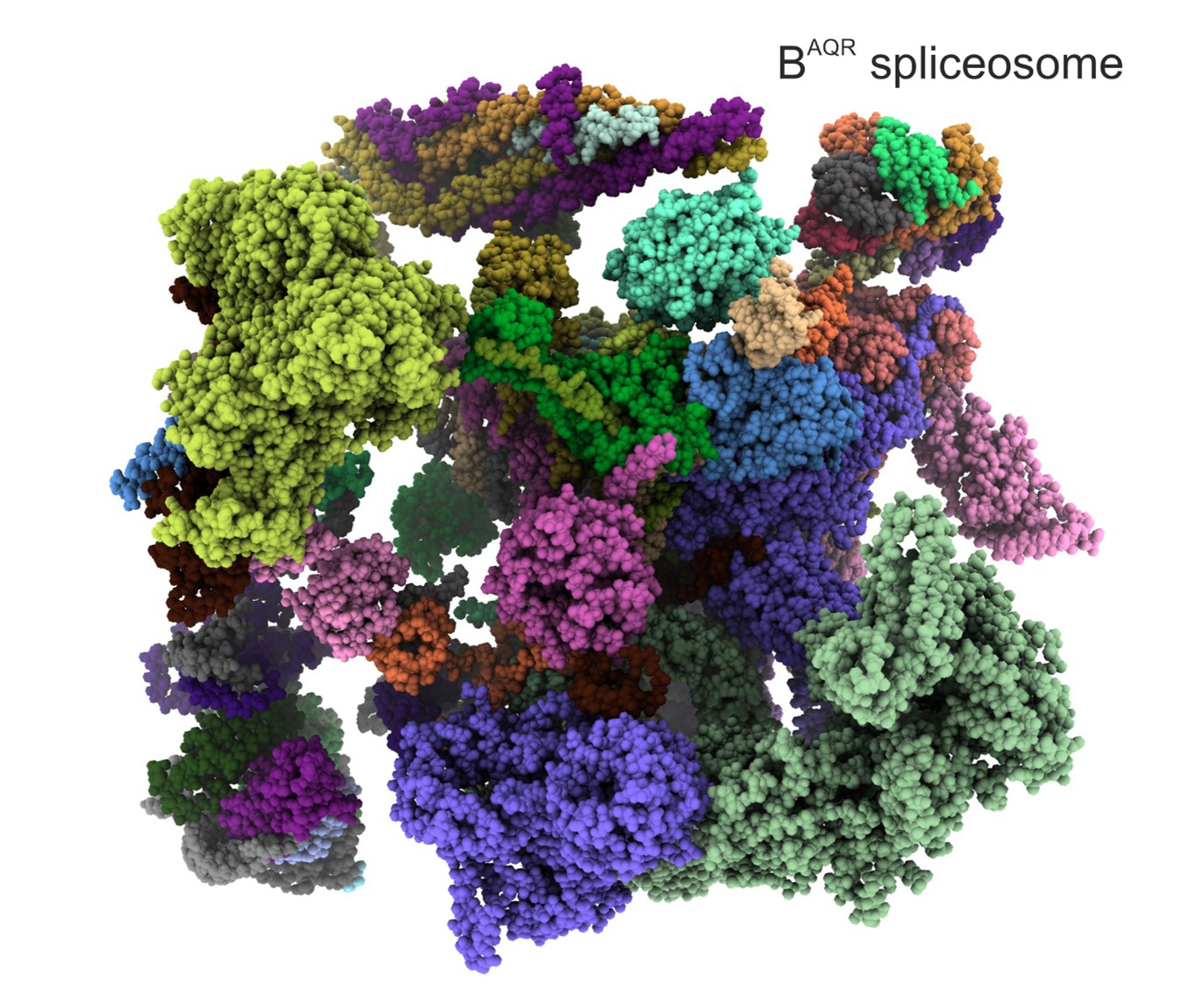
Identification of a spliceosome stalled halfway through the catalytic activation (Schmitzova et al., Nature, 2023).
Mechanism of the helicase PRP2 in splicing
The DEAH helicases are special as they can grasp an RNA strand and translocate along nucleotide by nucleotide while hydrolyzing ATP molecules. Four DEAH helicases are essential for splicing, although their mechanism of action in splicing remained unclear, as they were observed at fixed positions on the periphery of the spliceosomes. The BAQR spliceosome, found halfway through the catalytic activation, has finally shown how a DEAH helicase, PRP2, translocates about 19 nucleotides from the periphery towards the core of the spliceosome while remodelling the RNA-protein contacts, changing conformations, dissociating and recruiting proteins.
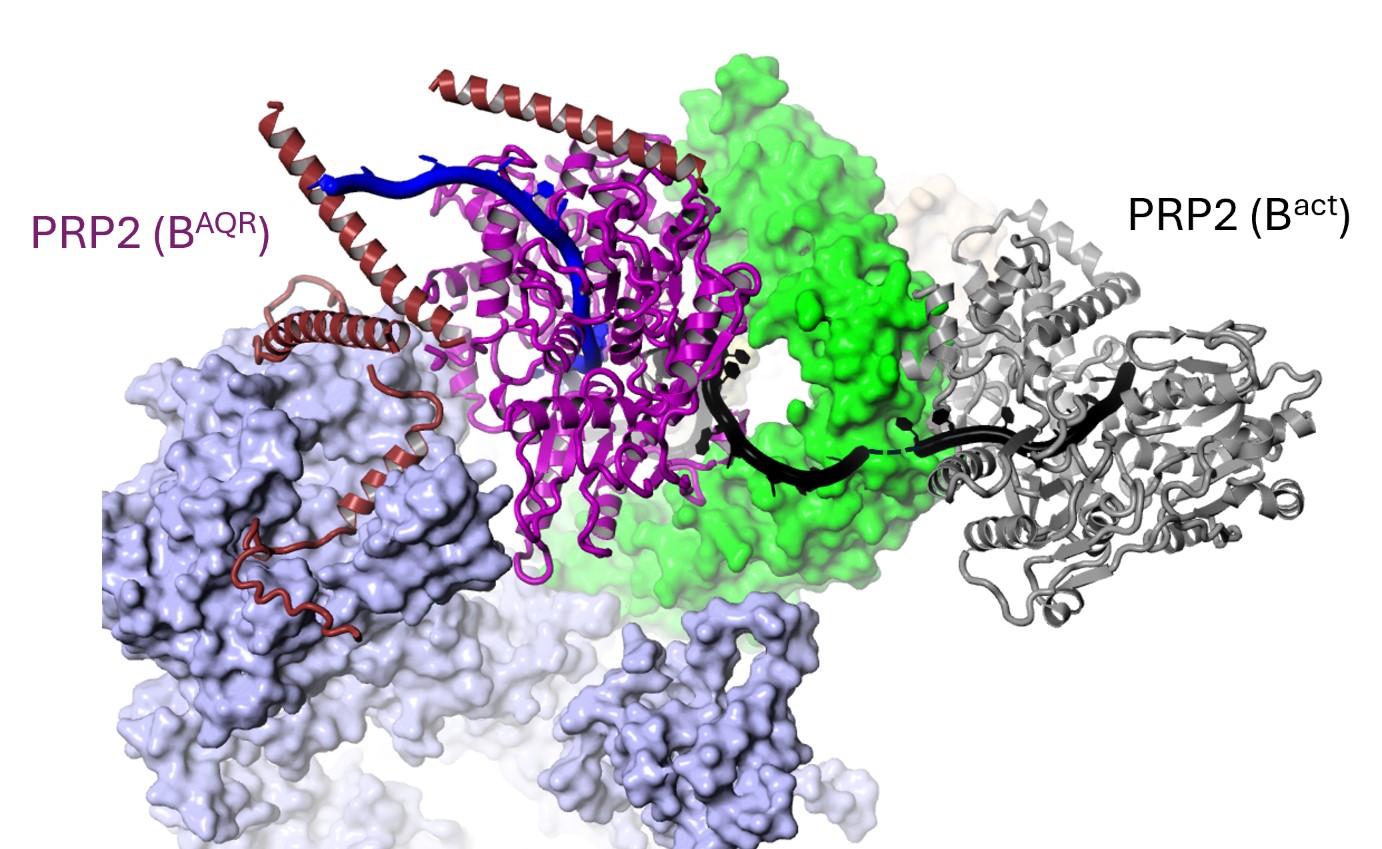
The helicase PRP2 translocates 19 nucleotides at the transition from Bact to BAQR (Schmitzova et al., Nature, 2023).
General mechanism of intron recognition during spliceosome assembly
This indicates that early prespliceosomes (E complexes) select introns through a toehold-mediated strand displacement mechanism. Strand displacement is primarily known from the DNA field, on processes like the DNA strand exchange, and in applied DNA nanotechnologies.
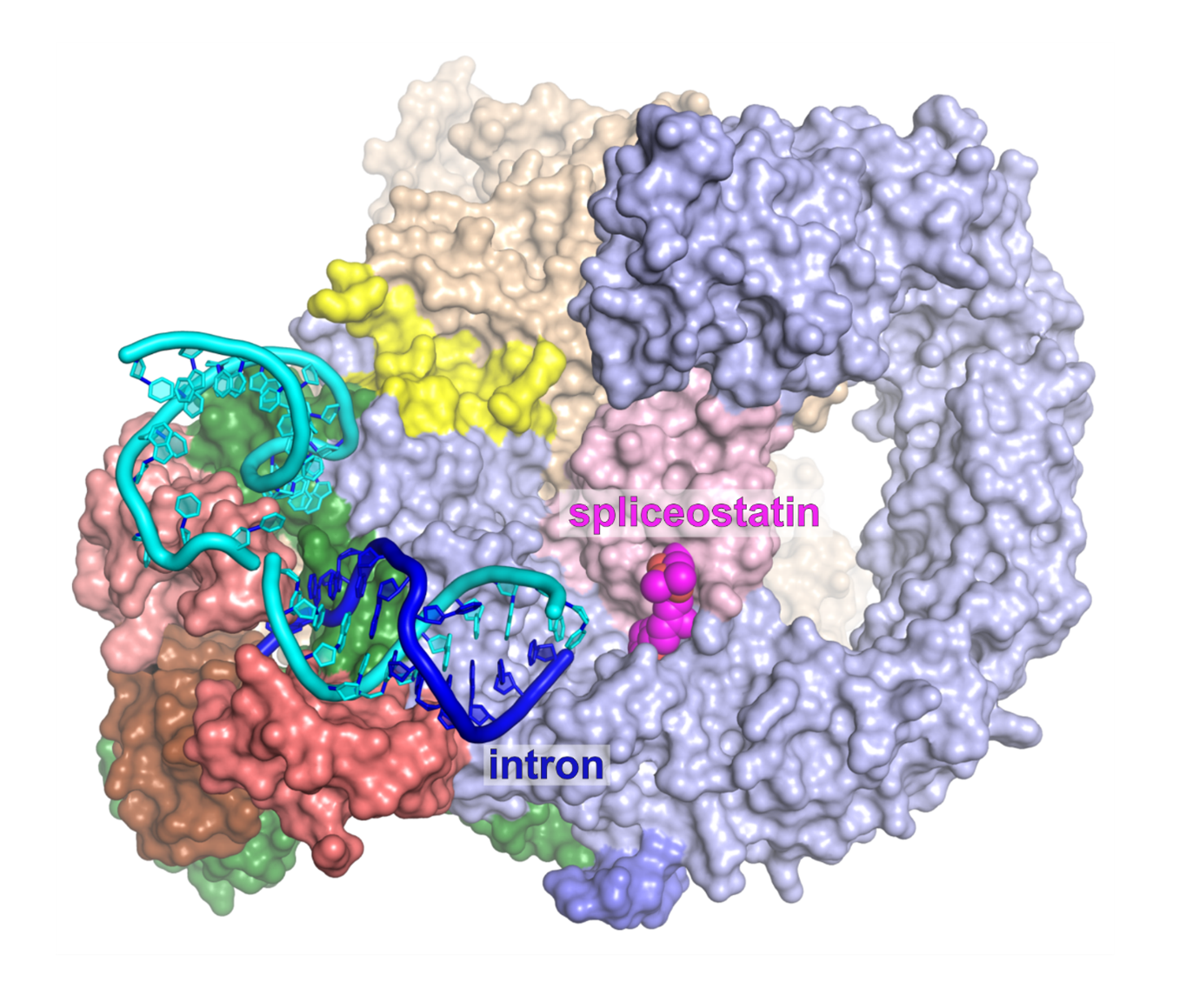
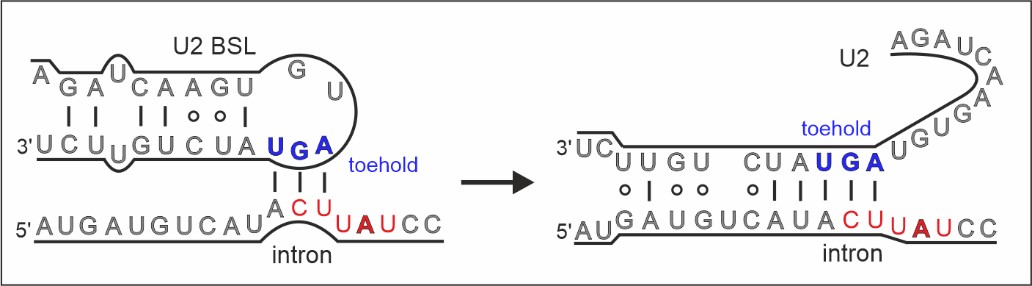
The structure of the exon-definition complex core stalled with spliceostatin (above) and the mechanism of intron recognition by strand displacement (below; Cretu et al., Nat. Comm., 2021).
General mechanism of splicing modulation by SF3B ligands
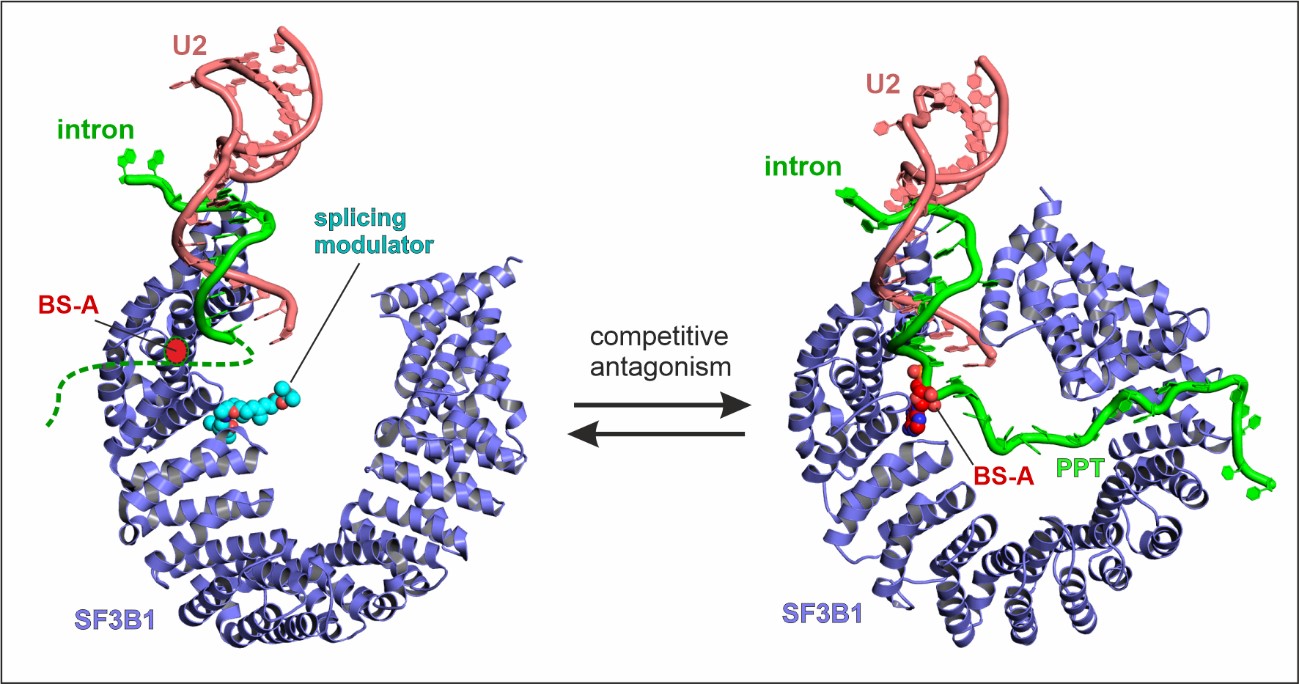
Splicing modulators that bind SF3B1 act as competitive antagonists of the branch site, in a manner dependent on the intron’s sequence (Cretu et al., Mol. Cell, 2018; Nat. Comm., 2021)
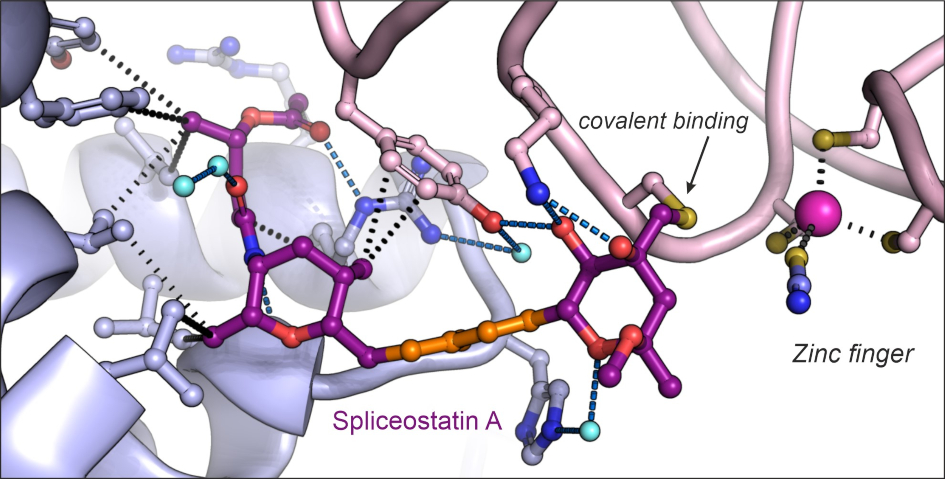
Mechanism of covalent coupling of spliceostatin and sudemycine to a zinc finger of SF3B
Our crystal structures have revealed that the long-studied compounds from the spliceostatin family and their relatives, sudemycins, bind prespliceosomes by covalent coupling to a zinc finger of the PHF5A protein. The central conjugated diene of the compounds acts as a spacer for the warhead epoxy group that reacts with a thiol of the cysteine.
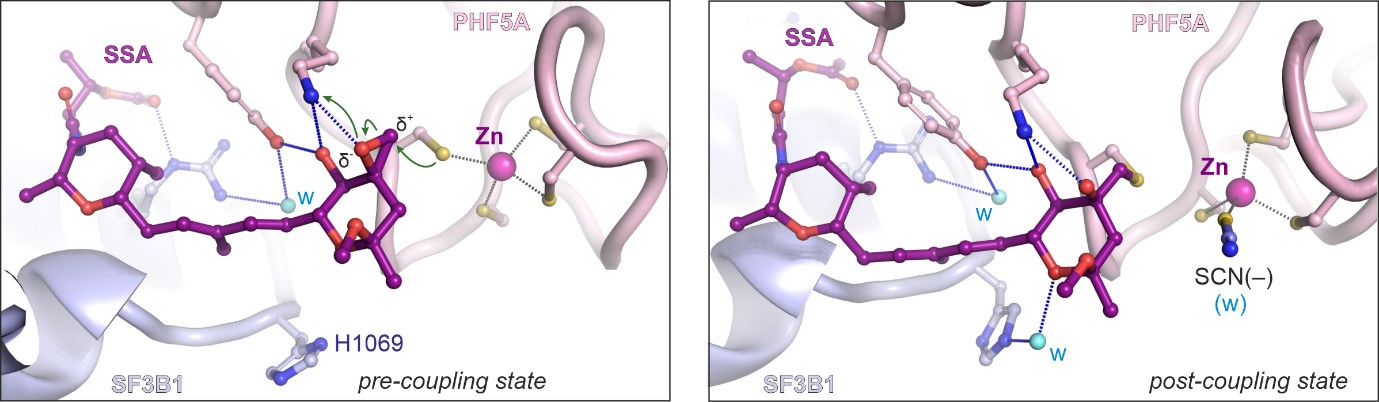
Spliceostatin and sudemycins bind spliceosomes by covalent coupling to a zing finger from the SF3B subunit PHF5A.
The NineTeen Complex (NTC) promotes ubiquitination in DNA repair
As part of the heteromeric NineTeen Complex (NTC), Prp19 is a homotetramer complex that promotes ubiquitination during the formation of the spliceosomal tri-snRNP particle and DNA damage response. We showed by X-ray crystallography and functional analyses in vitro and in vivo that Prp19 is an autoinhibited E3 ubiquitin ligase that becomes active only in stable association with three other NTC components that exert specific effects on Prp19's conformation. (De Moura et al., Mol. Cell, 2018).
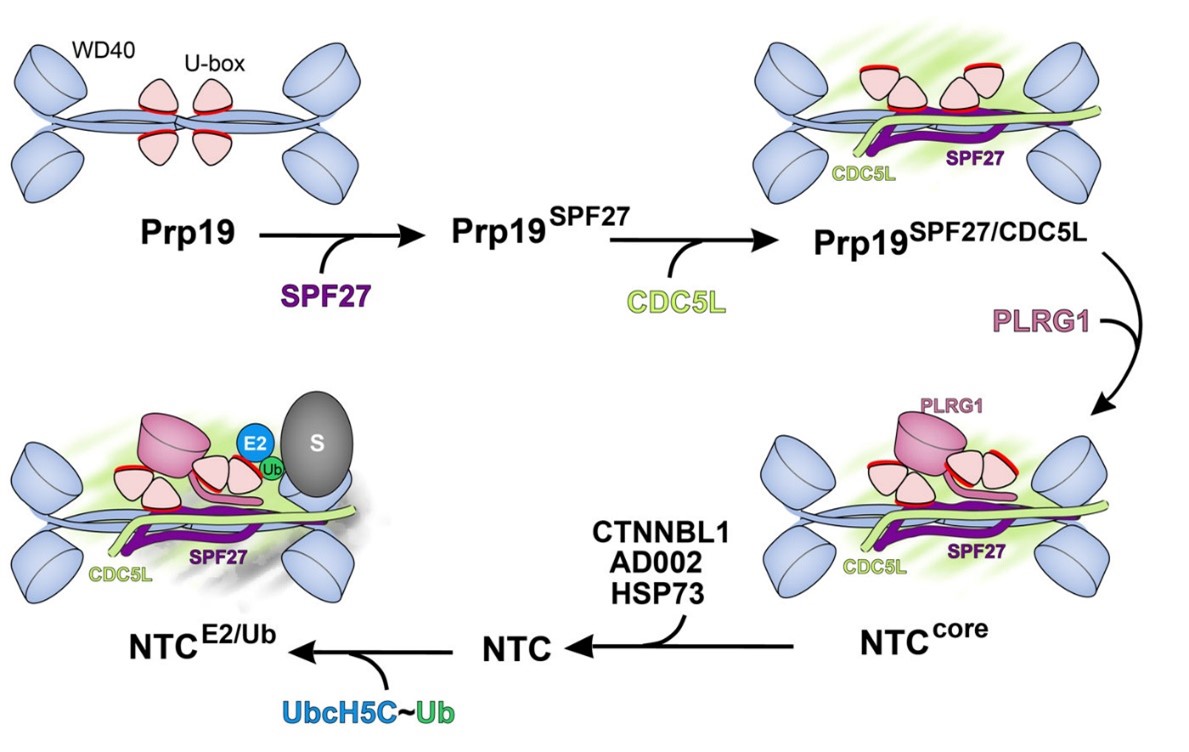
PRP19 is an autoinhibited E3 ligase activated by stepwise assembly of three splicing factors, together forming the NTC’s core
The Intron-binding complex (IBC) enables recruitment of the splicing-essential helicase Aquarius (AQR, IBP160) to the human spliceosomes
The human helicase Aquarius induces an additional ATP-dependent remodeling of the spliceosome. We determined the structure of Aquarius and showed that this helicase is recruited to the spliceosome as part of the pentameric intron binding complex (IBC).
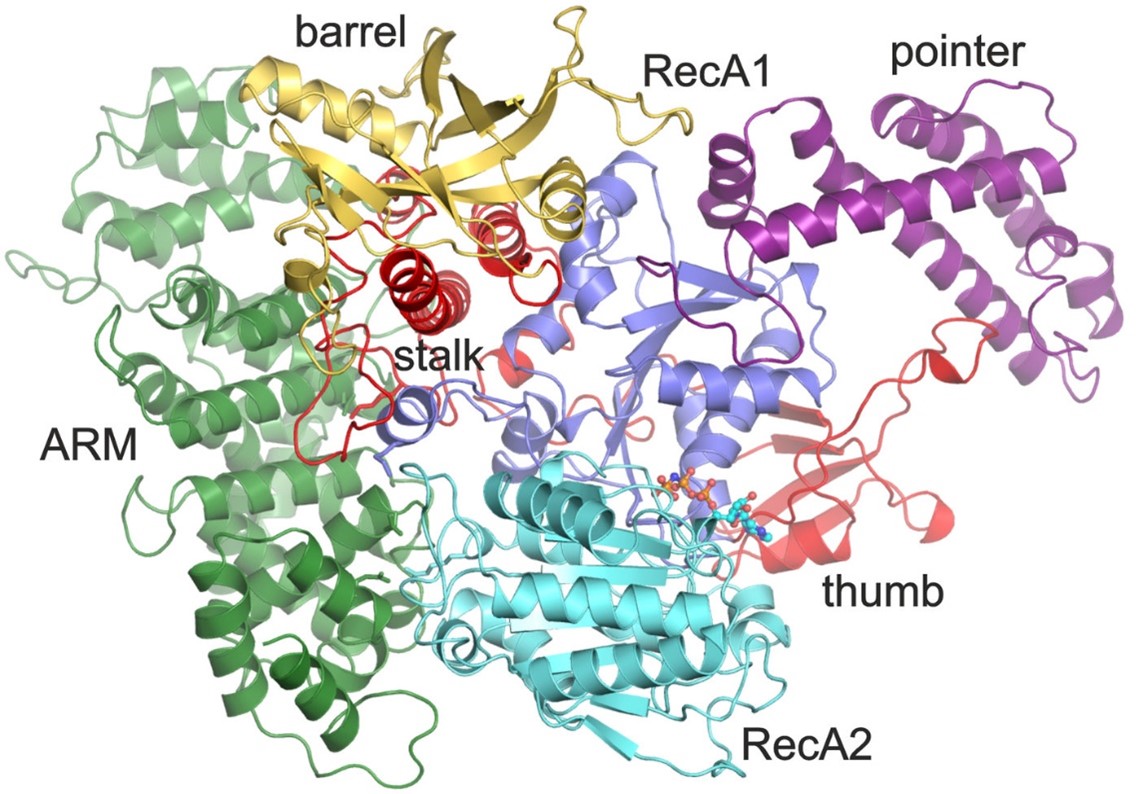
Crystal structure of the RNA helicase Aquarius
Crystal structure of a DNA catalyst
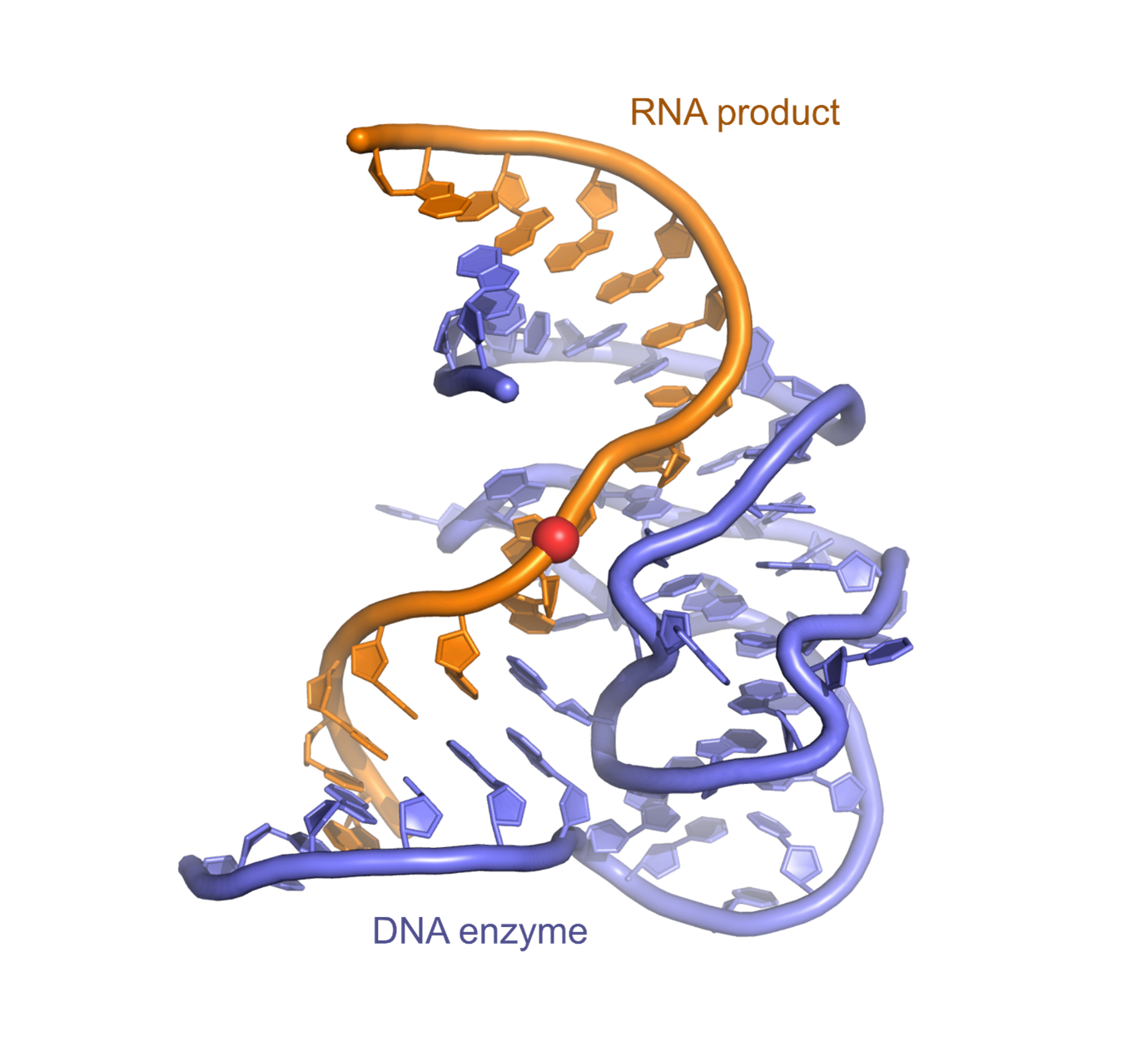
Crystal structure of the deoxyribozyme 9DB1 in complex with the RNA product
Professor Vlad Pena
Group Leader:
Mechanisms and Regulation of pre-mRNA Splicing
Professor Vlad Pena uses biochemistry, electron cryo-microscopy and X-ray crystallography to understand splicing regulation and the connections between splicing and other gene expression processes. He is a Professor of Structural Biology and Gene Expression at the ICR.
Researchers in this group
 .
.
Email: [email protected]
Location: Chelsea
Imogen is a PhD student in the Division of Structural Biology focused on designing small molecule splicing modulators, in collaboration with the Centre for Cancer Drug Discovery.
 .
.
Tiffany is a postdoctoral fellow who works on SF3B functions relevant to cancer.
 .
.
Email: [email protected]
Location: Chelsea
Judit is a higher scientific officer studying the regulation of RNA processing.
 .
.
Email: [email protected]
Gus explores the modulation of gene expression by oncolytic viruses.
 .
.
Email: [email protected]
Location: Chelsea
Csaba is a postdoctoral fellow working on splicing regulation.
 .
.
Hyunah is a postdoctoral fellow who studies the regulation of alternative splicing.
Professor Vlad Pena's group have written 24 publications
Most recent new publication 7/2023
See all their publicationsNews and discoveries from this group

 .
.