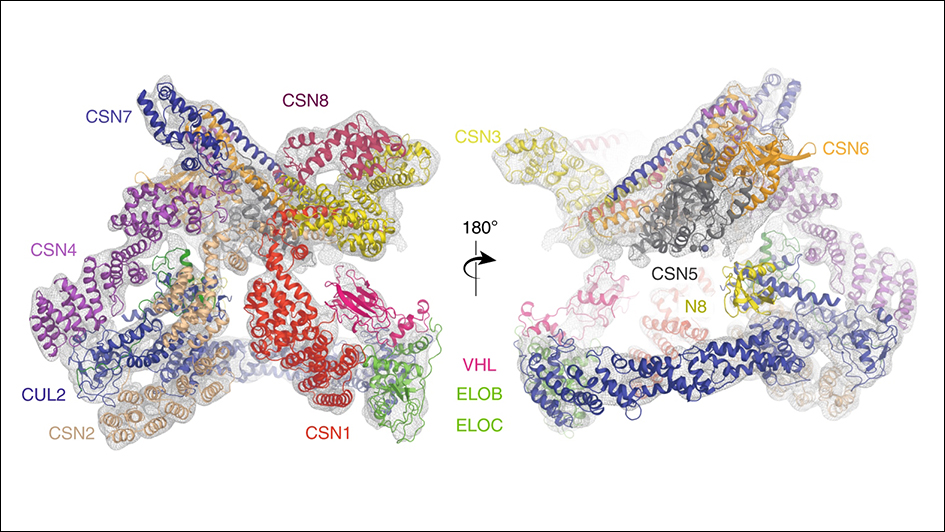
Image: Structures and interactions of the CSN–CRL2~N8 complex. Image also shows the molecular model of CSN–CRL2~N8 fitted into cryo-EM density (8.2 Å resolution) from front and back views.
Researchers have gained new understandings of two key complexes of cancer-related proteins by producing the most detailed ever maps of the structures they form when they come together.
The study reveals that the two protein complexes come together in cells in a series of steps that change how their individual component proteins are arranged together.
It is this change in the conformation of the component protein complexes which enables them to interact with yet more proteins that regulate cancer-causing processes.
The new insights into this system, published in the journal published in the journal Nature Communications, will help to direct other researchers in their efforts in cancer drug discovery – including in an emerging, cutting-edge field in which researchers are aiming to find ways to promote protein degradation using new therapies.
A possible target for cancer drugs
In the study, a team led by researchers at The Institute of Cancer Research, London, and King’s College London, used state-of-the-art equipment to map the structure of the assembly formed between two protein complexes – called Cullin-RING E3 Ligase 2 (CRL2) and the COP9 signalosome (CSN).
In cells, CSN binds to and deactivates CLR2, leading in turn to the activation of a third complex called HIF-1 alpha, which allows tumours to grow more effectively. The researchers pieced together a detailed ‘step-wise’ system explaining the mechanism of this system, which can ultimately drive cancer.
As well as shining new light on the role of these proteins in cells, and the part they play in cancer, the study is directly relevant to researchers who are currently studying CRL2 as a possible target for cancer drugs.
The complex has been proposed as a target for both small-molecule drugs, which could ‘lock’ into the protein to alter its function, and for an exciting new drug type called PROTACs, which aim to degrade tumour-driving proteins by recruiting E3 ligases like CRL2.
One of the key findings of the study was to clarify the role of an activating sub-unit of the CL2 complex called NEDD8, uncovering the existence of forms of CL2 which seem to have a biological role even without it.
Importantly, the scientists also brought together cryo-electron microscopy (cryo-EM) and two different types of mass spectrometry to make their discoveries. Their study provides a template for other researchers to marry these different techniques, gaining understandings of the structures and functions of proteins more efficiently than has previously been possible.
Let’s finish it: help us revolutionise cancer treatment. We aim to discover a new generation of cancer treatments so smart and targeted, that more patients will defeat their cancer and finish what they started.
Opening up new avenues of treatment
The study was supported by a range of funders including Cancer Research UK, The BBSRC, the Wellcome Trust and the Medical Research Council.
Scientists in the ICR’s Division of Structural Biology play an important part in our work to discover new targeted cancer drugs, mapping proteins of interest to the cancer research community and so opening up new avenues of treatment.
Using some of the most advanced technology available to structural biologists in the UK, they also uncover new information about the fundamental processes that underpin life itself.
Study co-leader Dr Edward Morris, Team Leader in Structural Electron Microscopy at the ICR, said:
“Our study used a range of structural biology techniques to generate detailed maps of both the Cullin-RING E3 Ligase 2 and the COP9 signalosome, both of which have a range of activities in cells and in combination have a role in some cancers.
“We hope our findings have an influence on the wider community of cancer researchers who are looking to discover innovative cancer drugs, including in the search for protein degradation therapies that could target E3 ligases like CR2.”
