Closed: Targeting SRC in G4-Medulloblastoma
Application closing date: 09/03/25
Project background
Medulloblastoma is the most common malignant brain tumour in children. It occurs in the cerebellum and accounts for approximately a quarter of all childhood brain tumours. It can be divided into 4 subtypes based on multi-omic profiling: WNT, SHH, Group 3 and Group 4.[1] Group 4 is the most common with a mixed prognosis for survival. Our collaborators at the Institut Curie, Paris and NCNP, Tokyo have shown that signalling through the non-receptor tyrosine kinase SRC is upregulated in Group 4 medulloblastoma and furthermore, that expression of SRC in combination with p53 inactivation leads to murine tumours that resemble group 4 medulloblastoma.[2]
This PhD project will seek to apply chemical biology to explore the role of SRC in medulloblastoma. The project will be multidisciplinary in nature involving medicinal chemistry, recombinant protein production, assay development, characterisation of protein-ligand interactions and structural biology. The aim will be to identify and make tool molecules that can target specific forms of SRC that will be further validated by our collaborators (the Ayrault’s Team, Institut Curie, Paris-FR).
The project will be based in the multi-disciplinary Centre for Cancer Drug Discovery (CCDD) at the Institute of Cancer Research (ICR), in the labs of Dr Gary Newton and Dr Rob van Montfort. The project will also be supported by Prof Paul Workman and Prof Louis Chesler.
The project is funded by the Cancer Research UK Children’s Brain Tumour Centre of Excellence (CRUK CBTCE), which is jointly led by Prof Richard Gilbertson (University of Cambridge) and Prof Paul Workman (ICR). The aim of the centre is to convene a critical mass of expert personnel, infrastructure and global collaborations in paediatric brain tumour biology, medicinal chemistry, pharmacology, and preclinical and clinical trials. Building on the success of our previous 6-year programme (https://www.crukchildrensbraintumourcentre.org ) we have recently received a 5-year renewal of funding from CRUK. The successful PhD candidate will benefit from being part of the CRUK CBTCE Early Career Research Network. This will offer opportunities to visit collaborating labs, participate in Symposia, Summer Schools and present at internal and external meetings.
Application deadline: 09/03/2025
Project aims
- Generate protein of full length SRC variants and specific domains and evaluate reported inhibitors
- Perform fragment screen against specific domains of interest; characterise and elaborate hits through application of medicinal chemistry, biophysical techniques and structural biology to generate more potent binders
- Establish assays for characterising the molecules that are generated
- Evaluate potential for generating compounds that can degrade SRC variants and establish whether it is possible to selectively degrade different forms of SRC
Further details & requirements
The project will be multidisciplinary in nature and will involve elements of medicinal chemistry, recombinant protein production, assay development, characterisation of protein-ligand interactions, structural biology, and cell biology. The aim will be to identify and make tool molecules that can target specific forms of SRC and, in combination with our collaborators, use these to explore the role of SRC in this important childhood cancer. The exact mix will depend upon the skill sets and preferences of the candidate as well as progress that is made with the different strands of research.
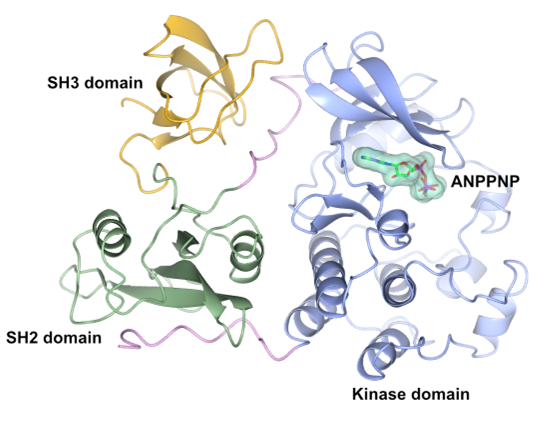
Figure 1. Crystal structure of human SRC kinase (residues 84-536, PDB ID 2SRC) with the kinase domain in bue and the SH2 and SH3 domains in green and yellow respectively. The bound ANPPNP molecule is shown in cylinder representation with a green semi-transparent surface overlayed.
Stage 1 – Generating and characterising recombinant SRC protein variants and characterising the binding of reported SRC inhibitors
SRC consist of several different domains, notably the kinase domain, the SH3 domain and the SH2 domain (Figure 1). There are numerous small molecules that have been reported to inhibit the kinase domain of SRC, some more selective than others.[3,4] The aim will be to generate a set of recombinant SRC protein samples to use for the biochemical, biophysical and structural characterisation of SRC inhibitors. The SRC variants will include full-length SRC as well as truncated variants comprising only the kinase domain and the respective SH2 and SH3 domains. The research will involve cloning of the required constructs, expression in either E. coli or insect cells expression systems followed by purification using the AKTA-purification systems of the HDSD team. Known SRC inhibitors and ligands will be either purchased or synthesised. To evaluate the potency of known SRC inhibitors, a biochemical ATP-turnover assay will be established for full-length SRC and/or the truncated kinase domain variant. The biochemical characterisation of known SRC inhibitors will be complemented by a biophysical and structural characterisation using the biophysical and structural approaches available within the HDSD team.
Stage 2 - Identifying and evaluating low molecular weight compounds or fragments that bind to specific domains and application of medicinal chemistry to generate more potent compounds
As mentioned above, the majority of reported SRC inhibitors inhibit the kinase domain of SRC but targeting the other domains with small molecules has been less well explored. We are particularly interested in targeting the SH3 domain as it has been reported to mediate the interaction of SRC with other proteins as well as influence the activation state of SRC.[5] Several peptides have been reported to bind to the SH3 domain of SRC which we intend to use as tool molecules to establish biophysical fragment screening approaches such as thermal shift assays (TSA), Surface plasmon resonance (SPR and/or ligand-observed NMR (LO-NMR). Using the SH3 domain protein synthesised in Stage 1 the PhD-student will carry out a fragment screen using a combination of these methods aiming to identify fragments that bind to the SH3 domain. It is anticipated that the student will further characterise these fragment hits using techniques such as surface plasmon resonance (SPR), fluorescence polarisation (FP) assays and X-ray crystallography before seeking to elaborate the hits using synthetic and medicinal chemistry, to generate compounds with a higher binding affinity.
Stage 3 – Identifying compounds that can degrade SRC and establishing whether it is possible to identify molecules that can target specific variants of SRC
- Generation of PROTACs from SH3 binders that have been identified in stage 2
- Generation of PROTACs from ligands that are known to bind to the kinase domain[6]
Compounds will be tested in cell-based assays established with our collaborators and in assays established in-house as part of the CRUK-CBTCE and CCDD.
Note: the ICR’s standard minimum entry requirement is a relevant undergraduate Honours degree (First or 2:1).
Pre-requisite qualifications of applicants:
BSc or MSc in a discipline related to chemistry, medicinal chemistry, chemical biology or life sciences. Chemistry applicant should have experience in Organic Synthesis, and Life Sciences applicants must have a basic knowledge of Molecular Biology. An interest in recombinant protein production and assay development will be an advantage.
Intended learning outcomes:
- An understanding of the role SRC in medulloblastoma
- Develop skills in recombinant protein production, biochemical and biophysical assay development, and structural biology applied to drug discovery
- Develop skills in chemical biology, organic chemistry and medicinal chemistry
- Develop the ability to test self-driven hypotheses, critically appraise relevant literature and design experiments independently using robust methodology
- Produce research outputs including project reports, conference abstracts, presentations at key meetings