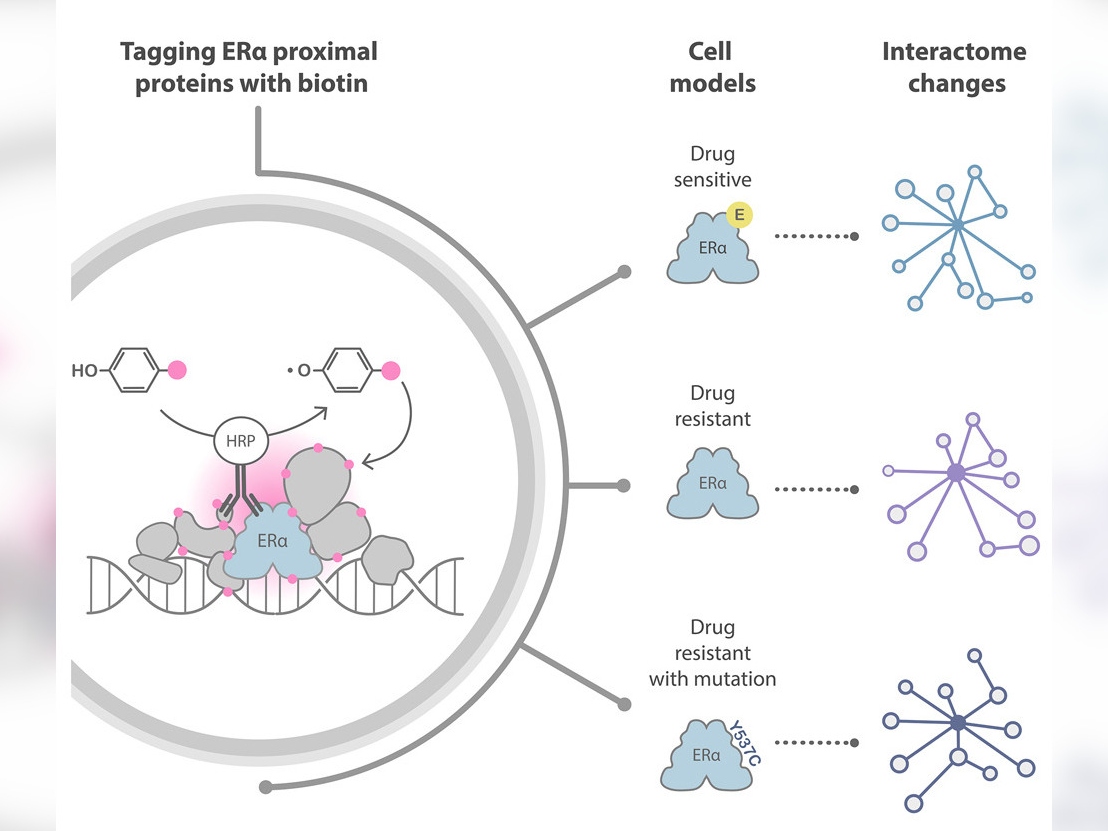
Functional Proteomics Group
Professor Jyoti Choudhary's group explores the molecular phenotype of cancer studying protein-protein interactions, signalling, post-translational modifications and protein expression.
Research, projects and publications in this group
We are interested in solving questions about the biology underlying cancerous disease by studying molecular processes and pathways. To do this, we use a range of techniques including mass spectrometry, biochemistry, molecular biology and informatics, to study protein function, proteome composition and organisation.
Professor Jyoti Choudhary
Group Leader:
Functional Proteomics
Professor Jyoti Choudhary's research focuses on the development and implementation of novel mass spectrometry and data analysis approaches for proteome discovery. She is Head of the Proteomics Core Facility and Career Faculty Leader of the Functional Proteomics Research Group.
Researchers in this group
 .
.
OrcID: 0000-0003-1478-3985
Phone: +44 20 7153 5119
Email: [email protected]
Also on: mrgraeme , https://twitter.com/mrgraeme
Location: Chelsea
My research uses the graph topology features of genomics and proteomics data to highlight previously undiscovered interactions, functional modules and pathway changes. I use a range of machine learning and explorative statistics techniques. I completed my PhD as part of the Genome Damage and Stability Center (GDSC) at the University of Sussex. My background is in Artificial Intelligence and Computer Science.
 .
.
My research is focused on deep profiling of protein expression, post translational modifications and protein interactions in cancer by mass spectrometry. I am interested in the integration of multi-omics approaches to better understand how genomic alterations affect the function of the proteome with implications in patient classification and response to treatment.”
Professor Jyoti Choudhary's group have written 180 publications
Most recent new publication 11/2024
See all their publicationsWe develop advanced mass spectrometry and novel proteomics techniques to further an understanding of cancer biology. Taking a multi-omics approach, we examine both the genome and the proteome (entire set of expressed proteins) to investigate the heterogeneity of tumours.
Through its research and collaborations our group explores protein-protein interactions, cell signalling and protein expression to investigate cancer in a range of human tissues, cell lines, and model organisms.
Cancer Proteomes
Assessing the impact of genomic alterations on protein networks is fundamental in identifying the mechanisms that shape cancer heterogeneity. We use isobaric labelling to characterize the proteomic landscapes of various cancers and decipher the functional consequences of somatic genomic variants. Using robust quantification of proteins and phosphopeptides enables the de novo construction of a functional protein correlation network, which ultimately exposed the collateral effects of mutations on protein complexes.
We also leverage the quantified proteome to build predictive models of drug response in cancer. Overall, we take a deep integrative view of the functional network and the molecular structure underlying the heterogeneity of cancer.
Proteogenomics and Personal Proteomics
As part of the GENCODE consortium we use proteomics data to assist in the complete annotation of human and mouse protein coding genes. Using integrative pipelines we collate multi-omics data to validate the existence and translation of protein isoforms. Targeted experimental approaches are used to discover and validate novel protein coding genes, and develop tools and methods for the analysis of paired transcriptomic and proteomic data. Additionally, we conduct experiments across multiple individuals to investigate personal variation and the influence of genome on the proteome.
Proteome Characterisation and Quantification Method Development
We are constantly developing novel mass spectrometry and informatics techniques to identify and quantify proteins and their modifications. These include profiling the changes in protein expression in cancerous tissues, analysing protein localisation to subcellular organelles, examining protein synthesis and turn-over and determining the absolute quantification of protein species.
 .
.