Last month, DeepMind published the much anticipated, detailed methodology underlying the latest version of AlphaFold – the UK-based science company’s powerful AI system that blew away its rivals in the latest major competition to predict the 3D structure of proteins.
AlphaFold’s machine learning methodology has been applied to predict structures for almost 99% of human proteins which have now been made publicly available. In this long read, I reflect on the significance of these developments for fundamental research and drug discovery.
I wrote this as the ICR celebrates the 10th anniversary of its AI-enabled drug discovery knowledgebase canSAR – which features multiple approaches to predicting ‘druggability’ as an aid to selecting drug targets and accelerating drug discovery.
The coronavirus pandemic has, understandably, soaked up a lot of bandwidth when it comes to science news – but this particular non-Covid science story was able to cut through and hit the headlines in the UK and around the world.
On 30 November 2020 it was announced that DeepMind – a subsidiary of Google’s parent company Alphabet focusing on artificial intelligence – had made what was hailed as a huge leap towards solving one of biology’s greatest remaining challenges: the ability to predict the correct, three-dimensional structures of proteins based on their constituent, one-dimensional amino acid sequences.
The announcement attracted huge interest, but the expert community has been waiting for the peer-reviewed science publication. The AI methodology has now been published in the leading journal Nature and this was followed rapidly by a second Nature paper from DeepMind and collaborators at the European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), which reports the application of the most recent AlphaFold machine learning system to predict the 3D structures at scale for almost the entire human proteome – 98.5% of human proteins.
Founded in the UK in 2010, a statement on DeepMind’s website says: ‘Our long-term aim is to solve intelligence, developing more general and capable problem-solving systems, known as artificial general intelligence (AGI).’
DeepMind made an initial impact by using its AI methods to beat a professional Go game champion and by learning to play from scratch 49 different Atari games – based only on the pixels and the score on the screen.
DeepMind’s intention was to use these gaming challenges as a basis upon which to develop solutions to major, real world problems.
Indeed DeepMind’s AI technology has been applied successfully in the biomedical field – for example in identifying early signs of retinal disease from eye scans and the detection of breast cancer from mammography scans.
The company’s strategy is to use advanced AI methodology known as machine learning and neural networks to power its predictions.
As a further real world, major biomedical challenge, DeepMind decided to focus its attention on making much-needed improvements in our ability to predict accurate 3D structures of proteins based on the linear polymeric sequence of amino acids – which are encoded in our genes and arranged like the beads on a string.
The importance of protein folding
Before looking at what was achieved by DeepMind’s latest version of AlphaFold, we need to consider why it is that being able to predict the different 3D origami structures into which different proteins fold up is such a big deal.
This is because proteins perform the biological functions that are required for life – whether, for example, that be catalysing a biochemical reaction, recognising an infection, or enabling muscles to contract. And knowing the 3D shape of proteins is important because it is a foundational principal in biology that structure determines function, in the same way that the design and shape of a piece of furniture – say a chair, table or bed – dictates how it is used.
Overall, proteins fall into two main structural classes: globular proteins, which are compact, spherical and soluble, and others that are elongated, fibrous and insoluble.
Within these broad classes, individual proteins fold up into a unique shape, the exact details of which define the jobs they carry out. So determining the precise 3D structure of a given protein is a crucial part of understanding how it performs its function in the cell and in the body – and therefore is vital to understanding the molecular basis of life itself.
Truly a grand challenge!
Furthermore, as I address later, in addition to the importance of being able to predict protein structure for our fundamental molecular comprehension of life, there are also huge practical benefits – for example in understanding and treating disease and in the design of proteins for practical use in the biotechnology industry.
So no pressure there either!
The historical context
It’s important, too, to view the protein folding grand challenge in its historical context.
The so-called ‘Central Dogma of molecular biology’ was originally hypothesised in a conference lecture in 1957 by the British physicist-turned-biologist Francis Crick. In that talk, Crick famously used the memorable depiction ‘DNA → RNA → protein’, where the arrows indicate the ‘flow of information’. Crick of course later won the Nobel Prize with Jim Watson (and also Maurice Wilkins with a major contribution from the by then sadly deceased Rosalind Franklin) for their research published in 1953 in which they solved the double-helix structure of DNA – and which revealed the explanation for how genetic information encoded by the sequence of base pairs in the double helix can be precisely copied.
Also importantly, Crick’s extraordinary insight into the fundamental molecular information flow of life proposed correctly in broad terms how the 1D linear sequence of bases in DNA – making up the genetic code – could be translated, via the corresponding 1D messenger RNA sequences, into the 1D sequence of amino acid building blocks that make up the polypeptide chains of proteins.
However, although brilliant, the Central Dogma did not explain at all how the linear amino acid chains are folded up into the correct twists and turns of the proteins that make up the 3D molecular machines of life.
Twelve years later, in another important conference lecture delivered in 1969, the American molecular biologist Cyrus Levinthal estimated that the number of theoretically possible configurations that a representative polypeptide chain might adopt could be as high as the mind-blowing number of 10300 – meaning that the protein would take longer to fold correctly than the age of the then-known universe. This is in stark contrast to the observed ability of proteins to achieve their correct 3D shape in as little time as a few seconds. The conundrum became known as the Levinthal Paradox of protein folding.
A further three years later, in 1972, the US biochemist Christian Afinson won the Nobel Prize for the ‘thermodynamic hypothesis’ – based on his studies of the protein called ribonuclease, and ‘in particular the relationship between the amino acid sequence and the biologically active conformation’.
Afinson concluded his Nobel prize lecture paper with a look into the future when it would be possible predict how the 3D shape of a protein structure could relate to its corresponding genetic code and amino acid sequence, saying:
‘Empirical considerations of the large amount of data now available on correlations between sequence and three dimensional structure, together with an increasing sophistication in the theoretical treatment of the energetics of polypeptide chain folding, are beginning to make more realistic the idea of the a priori prediction of protein conformation. It is certain that major advances in the understanding of cellular organization, and of the causes and control of abnormalities in such organization, will occur when we can predict, in advance, the three dimensional, phenotypic consequences of a genetic message.’
The Critical Assessment of protein Structure Prediction (CASP) ‘competition’
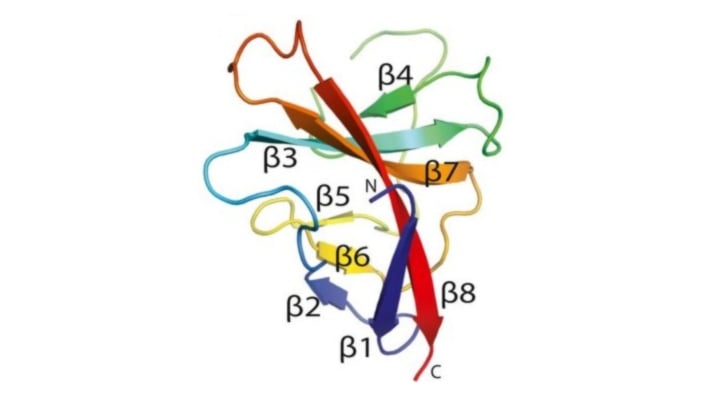
Image: Cartoon representation of the SARS-CoV-2 ORF8 monomer. β-strands are labelled β1 to β8. Credit: Flower, T.G. et al. Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein. PNAS (2021).
Despite gradual progress being made, in 1994 the frustratingly limited ability to correctly predict the 3D conformation of proteins from the constituent amino acid sequence led a group of structural biologists to set up the Critical Assessment of protein Structure Prediction (CASP) ‘competition’. Note that the organisers don’t like the term ‘competition’ and prefer ‘experiment’, but to outsiders it certainly looks like a competition!
The aim of this biennial exercise is to accelerate progress. Every two years around a hundred research teams strive to see how accurately their computational methods can predict the 3D structures of proteins for which the solutions have been revealed by painstaking empirical structure determination – typically done by X-ray crystallography or cryo-electron microscopy – but not yet publicly released.
Because the entrants in the protein structure prediction Olympics are asked to predict the structures of a hundred proteins ‘blind’, CASP really does challenge each team’s ability to make accurate estimates.
To provide a bit of technical detail, the accuracy of protein folding predictions is measured by the so-called global distance test or GDT. This compares the predicted 3D structure of a given protein with the experimentally determined conformation – which represents the so-called ‘ground truth.’
In the latest round – CASP 14, which was run over several months last year – DeepMind’s new AlphaFold AI technology (referred to as AlphaFold2) blew the other entrants away by achieving a median GDT score of 92.4 across all the proteins to be solved in the competition – with 100 being the highest possible score.
This would effectively represent 100% perfect accuracy, with all the atoms in the amino acids being in the precisely correct position. Anything over 90 is comparable with the results of laborious experimental determinations of protein structures and is considered as a correct solution that scientists can rely on with confidence.
For the hardest category of all – known as free-modelling, where the teams are asked to predict protein structures that do not have any comparable related protein structures available as template guides – the new AlphaFold2 technology achieved the remarkable median GDT score of 87.0.
Overall, the DeepMind team’s predictions had an average error (calculated technically as the root-mean-square deviation or RMSD) of approximately 1.6 Ångströms – where one Ångström is equal to 1 x 10-10 metres or 0.0000000001 metre. This means that the error in prediction is as small as the estimated width of a carbon atom (1.4 Ångströms).
So not too bad at all!
To emphasise just how much progress had been made, compare the above-mentioned AlphaFold2 median GDT score of 92.4 with the score achieved by DeepMind’s previous version of AlphaFold, which was around 70-75. This earlier version won the CASP 13 exercise in 2018 – representing the first major impact of AI on protein structure prediction. Before that the GDT scores achieved in several previous CASP competitions were down around the 60 mark.
Importantly, the new AlphaFold2 AI predictions in CASP 14 worked well across a wide range of different types of protein, which is obviously important for the general application of the methodology.
An interesting and topical protein in the competition was the SARS-CoV-2 virus protein ORF8, the structure of which was subsequently published.
ORF8 is a rapidly evolving coronavirus protein that has been implicated in immune evasion. Not only did AlphaFold accurately forecast the conformation of the components of the 3D shape known as antiparallel β-sheets – a common structural motif found in proteins – but even more impressively, the new methodology apparently also got it right in forecasting the spatial location of the loops that connect the β-sheets together. Such loops are notoriously difficult challenges in structure prediction.
Positive reception
The response of experts in the field of protein folding was widely very positive, although it was acknowledged that a full analysis of the new AlphaFold2 AI approach must await formal peer review and publication – as duly occurred with the earlier version of AlphaFold – and some commentators pointed out the current inevitable limitations.
A news piece in the leading journal Science quoted the expert computational protein scientist Janet Thornton as saying, ‘What the DeepMind team has managed to achieve is fantastic and will change the future of structural biology and protein research.’ The Science report also quotes the Nobel Prize-winning structural biologist Venki Ramakrishnan, who described the work as ‘a stunning advance on the protein folding problem’.
Similarly, in a Nature news piece the computational biologist and co-founder of CASP John Moult said: ‘This is a big deal. In some sense the problem is solved.’ And the same news article also quotes Andrei Lupas, who studies protein evolution, as saying: ‘This will change everything’. Not surprisingly Lupas was impressed that AlphaFold enabled him to determine in half an hour the structure of a protein that he’d failed to solve for 10 years!
More detailed commentaries last November aimed at experts in protein informatics can be found on the Oxford Protein Informatics Group's blog and Mohammed AlQuraishi's blog.
How was it done?
In terms of how the AlphaFold2 breakthrough was actually achieved, last year DeepMind provided some initial information. The Science news piece paraphrases John Jumper from the AlphaFold team as saying that they combined deep learning with an ‘attention algorithm’ that mimics how humans assemble a jigsaw puzzle.
The team used a computer network built around 128 machine learning processors to train the algorithm on the 170,000 experimentally determined protein structures in Protein Data Base or PDB. DeepMind also said that the AlphaFold team used large databases containing protein sequences of unknown structure.
What was also clear is that the AlphaFold2 predictions would have required an awesome amount of compute power and benefited from intense team working at DeepMind.
The details are now made fully transparent in last month’s Nature publication. There, the authors state: ‘Underpinning the latest version of AlphaFold is a novel machine learning approach that incorporates physical and biological knowledge about protein structure, leveraging multi-sequence alignments, into the design of the deep learning algorithm.’
They also confirm that the neural network AlphaFold system they developed is a completely different model from their earlier approach used successfully in the previous CASP event.
In addition, the authors comment: ‘AlphaFold greatly improves the accuracy of structure prediction by incorporating novel neural network architectures and training procedures based on the evolutionary, physical and geometric constraints of protein structure. In particular, we demonstrate a new architecture to jointly embed multiple sequence alignments (MSAs) and pairwise features, a new output representation and associated loss which enable accurate end-to-end structure prediction, a new equivariant attention architecture, use of intermediate losses to achieve iterative refinement of predictions, masked MSA loss to jointly train with structure, learning from unlabelled protein sequences using self-distillation, and self-estimates of accuracy.’
For those needing it, further detail is provided in the text of the Nature publication and associated supplementary data. Also the source code for the AlphaFold2 model, trained weights and inference script are made freely available under an open-source license.
What are the benefits of AlphaFold’s predictive power?
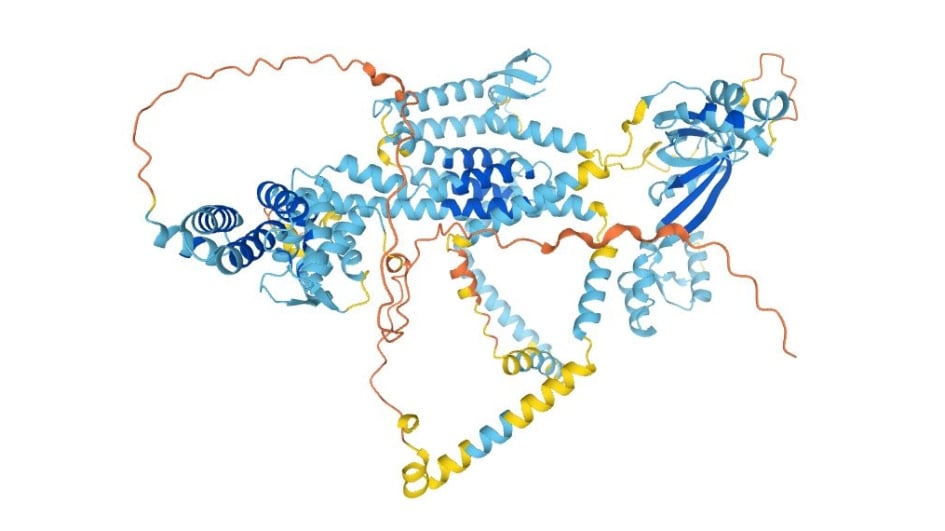
Image: AlphaFold structure prediction of the transmembrane endoplasmic reticulum protein wolframin. Credit: AlphaFold protein structure database. Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature (2021).
Access to the source code and other details not only allows independent validation of the AlphaFold2 predictions, but also researchers can now use the methodology to predict structures for their own proteins of interest.
An immediate and important benefit is that free access to AlphaFold’s protein structure predictions is enabled by DeepMind and the European Bioinformatics Institute (EMBL-EBI) through their AlphaFold Protein Structure Database. So researchers worldwide can use the predictive powers of DeepMind in their research.
Deep Mind is to be commended for this altruistic behaviour which will greatly widen the impact of its work.
As mentioned, the new AlphaFold machine learning system was used in the follow-up Nature paper to predict the 3D structures at scale for almost 99% of all 20,000 human proteins that are now in the database.
As well as the human proteome, the database also currently contains ~350,000 protein structures including most of the proteins present from 20 model organisms – including those from organisms commonly used in biomedical research, such as the bacterium E.coli, fruit fly, mouse, zebrafish and disease pathogens such as the malaria parasite and the tuberculosis bacterium.
In the second Nature article on AlphaFold2 published last month, the authors summarised both the overall performance of the new methodology and also the limits – as applied to 98.5% of the human proteins with full amino acid chain prediction.
In comparison to experimental determination over many decades that has provided structural coverage of only 17% of the total amino residues present in human protein sequences, AlphaFold now provides a dataset at scale that covers 58% of amino acid residues with a confident prediction – and within this dataset there is a subset for which 36% of all residues are predicted with very high confidence.
At the individual protein level, AlphaFold2 gives a confident prediction for at least three quarters of the amino acid sequence in the case of 44% of human proteins.
The authors say they have obtained high-quality structural predictions for human proteins ‘across a broad range of gene ontology (GO) terms, including pharmaceutically relevant classes such as enzymes and membranes’.
In the second Nature paper, the authors provide three interesting, illustrative examples of de novo predictions for the 3D structures of human proteins lacking significant templates, namely the transmembrane enzyme glucose-6-phosphatase, another enzyme diacylgycerol O-acetyltransferase 2, and the transmembrane endoplasmic reticulum protein wolframin for which mutations are linked to the rare genetic condition, Wolfram Syndrome. The three structures suggest biological hypotheses that can now be followed up – and this will be true for the predicted structures of very large numbers of other proteins.
There are however many regions across human proteins for which 3D structure cannot be predicted with confidence. Moreover, it is not surprising that the first author of the first of the two recent AlphaFold2 Nature publications John Jumper has commented: ‘It seems that in many cases where AlphaFold struggles it is because the protein itself is “disordered,” with no inherent structure in isolation.’
This is a significant limitation given that disordered regions comprise between 37 and 50% of human proteins – but it is noted that it is useful in itself to be able to predict which are the intrinsically disordered regions. In fact, predicting these regions computationally is the subject of a research community challenge in its own right.
It should be stressed that although prediction of full-length protein structure is always desirable, the ability to predict accurately the 3D shape of individual domains within proteins can be very valuable.
Also to be highlighted is the speed of the protein predictions. The first author of the second of the two recent AlphaFold Nature publications, Kathryn Tunyasuvunakool, has said that it took AlphaFold2 only around 48 hours to obtain the 350,000 protein predictions – an astonishing speed, especially compared with the typically laborious pace of experimental structure determination.
Overall the results provided show that excellent and exciting progress has been made in protein prediction using AlphaFold2 – while also revealing the scope for substantial further improvement.
The benefits are likely to be far reaching, both now and into the future.
The co-founder and CEO of DeepMind – and also last author on the two Nature papers – Demis Hassabis told Fortune magazine that the publication of AlphaFold’s structure predictions was his company’s ‘biggest contribution to science to date [and] an example of the benefits AI can bring to society’. See more comments from Hassabis in his recent blog post.
Fortune also quoted very positive views from Elizabeth Blackburn (University of California San Francisco), Paul Nurse (Francis Crick Institute) and Ewan Birney (EMBL-EBI). These biomedical research leaders and many others have praised AlphaFold’s accurate and large-scale protein prediction capability, with many seeing it as the biggest breakthrough since the determination of the human genome sequence around 20 years ago – and arguably similar in scale and impact.
A Nature editorial last month reported a consensus view that ‘it’s too early to predict exactly what impact the application of AI in the life sciences will have, except that any impact will be transformative.’
What is the likely impact on drug discovery?
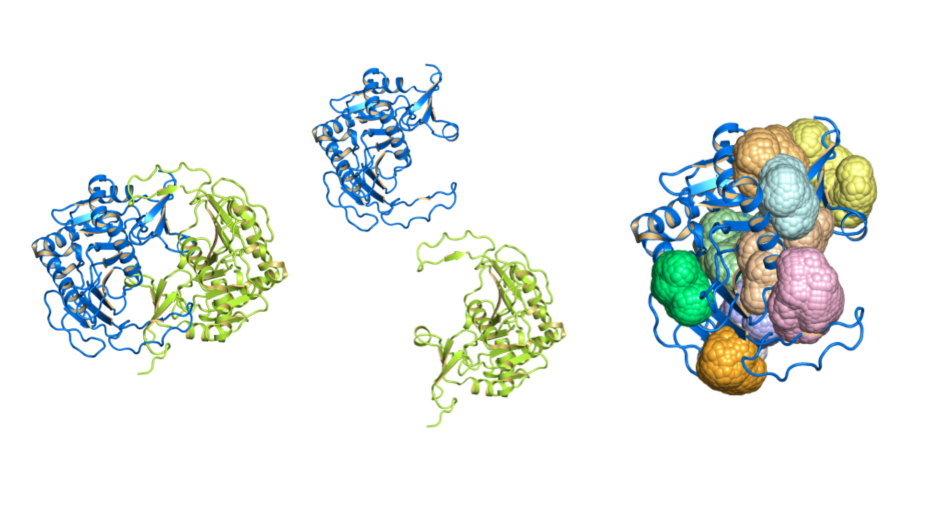
Image: 3D protein structures, chains and cavities visualised by canSAR. Credit: Patrizio Di Micco, ICR
There is no doubt that AlphaFold’s AI-powered protein prediction technology will greatly benefit fundamental research (understanding the structure and function of life), the biotechnology industry (engineering proteins as molecular machines and foods) and of course – as has been much promoted if not hyped in the coverage – the discovery of new drugs.
Most of the new drugs that we discover today for cancer and other diseases exert their effects by targeting particular proteins in the body. Ideally, we wish to design small-molecule drugs to bind very precisely to a tiny region of the overall target protein so as to alter its function. Medicinal chemists will always prefer to have an accurate 3D protein structure so they can apply structure-based drug design – thereby helping them to achieve as ‘perfect fit’ as possible alongside multiple other optimised properties.
And the 3D protein structure is important even before the drug discovery phase begins – by helping the drug discovery team to assess the druggability of the target protein – the extent to which it has pockets or grooves that allow small-molecule compounds to bind.
This allows researchers to understand which targets are relatively straightforward to drug and which ones will represent a major challenge. Such information can be extremely useful in prioritising which protein targets to pursue and the approach to be taken.
Here at The Institute of Cancer Research, London, we developed the canSAR knowledgebase which integrates massive multidisciplinary data to enable objective target assessment and prioritisation and already incorporates AI and machine learning techniques to predict druggability. We not only use canSAR in our own drug discovery research at the ICR, but we also made it freely available for use by researchers worldwide. As mentioned earlier, we have recently celebrated canSAR’s 10th anniversary.
There was certainly a buzz among my drug discovery colleagues at the ICR as we read the news last November about the AlphaFold2 breakthrough and discussed the potential impact on drug discovery. Our Head of Data Science, Professor Bissan Al-Lazikani who created canSAR, was in fact – early in her career – part of the team that won CASP4 in 2000.
Bissan has written in the Spectator magazine about what she feels this discovery could mean for her field – concluding: ‘If we can effectively harness Deepmind’s technology, we will gain a much better understanding of all the proteins and mutations that cause cancers. It will help us to accurately design and discover better, safer drugs that could successfully treat or cure countless people.’
In the canSAR team that I am part of, we are currently evaluating for ourselves the impact of AlphaFold2 and incorporating the predicted structure models into the knowledgebase, starting with the human proteins.
But how much does knowledge or accurate prediction of protein structure speed up drug discovery – from target selection to clinical candidate and approved drug?
I have been personally involved in multiple drug discovery projects at the ICR in which exploiting the 3D structure of the target protein has played a major role. These include the targets Hsp90 in collaboration with Professor Laurence Pearl and AKT/PKB with Professor David Barford. In their different ways, both projects benefited very much from the availability of the protein structure and both resulted in drugs entering clinical trial.
There is no doubt that the ability to predict accurate 3D protein structures is generally very valuable to drug discovery – and also, by the way, in the design of chemical probes to evaluate the role of the target in biology and disease pathology. Indeed the availability of a protein structure can in some cases make or break a drug discovery project – especially where a fragment-based approach is needed.
The AlphaFold team says that it is already collaborating with organisations such as Drugs for Neglected Diseases Initiative (DNDi) and other partners.
However, the response of some drug discovery professionals to the AlphaFold2 success has been more muted than some of the other commentary. This is exemplified by the views of the medicinal chemist and drug discovery blogger Derek Lowe – both last year and after the Nature publications last month.
While recognising the scientific advance, Derek points out to those uninitiated in the arts of drug discovery that: 1) predicted structures may be not be accurate enough and thus experimentally defined structures may still be needed; and 2) although actual or accurately predicted structures can accelerate the very early phase (let’s say saving an initial year or two) they do not greatly shorten the overall, say, 10 to 15 years that are needed from target selection (following validation) all the way to drug approval for widespread use.
What would represent an even bigger overall acceleration of drug discovery, as Derek rightly suggests, are solving tough problems such as ‘better prediction of useful drug targets, more translatable disease-predictive cell and animal models, and earlier assays that are more predictive of human toxicology.’ Also I would say the speeding up of clinical trials and the regulatory approval stages.
Personally, I think that (in addition to the major benefits in fundamental research) AlphaFold will have big impact on drug discovery – but there is no doubt that after the early stages where AlphaFold will have maximal effect, a lot remains to be done to discover and develop a drug where an accurate 3D protein structure has little to contribute.
However, as my colleagues and I have discussed, the application of big data and AI approaches more generally (ie beyond AlphaFold and protein structure determination) is clearly beginning to show benefit across the drug discovery and clinical continuum, with great potential to have further impact.
Has the Levinthal Paradox been resolved?
The answer to this is a definite no.
At least for now, what has been achieved by AlphaFold is not an answer to the question of the mechanisms by which proteins actually achieve the correct 3D fold, but rather only so far the ability to predict way more accurately than before the end result of protein folding.
The ultimate resolution of the Levinthal Paradox will require the physics and chemistry of the mechanisms by which proteins achieve their correct fold to be worked out. It’s possible, however, that this might be approached by ‘deconvoluting’ the AI system used by AlphaFold.
What is still left to be done?
Going beyond the 20,000 human proteins and more than 350,000 proteins overall for which AlphaFold2 has provided predicted 3D structures, DeepMind’s ambition is to increase this number to 130 million – approximately 50% of all known proteins – by the end of the year. That would be astonishingly rapid. And presumably the remaining extant proteome as well.
Beyond that and continuing to enhance the accuracy of protein structure prediction, what further challenges lie ahead for AlphaFold and other AI-driven approaches?
The use of AlphaFold2’s deep learning and neural networks approach to predict the following will be extremely important:
- Structures of intrinsically disordered regions
- Conformational changes and protein dynamics
- Effects of post-translational protein modifications
- Structures of multi-protein complexes
DeepMind and also others will continue to refine the deep learning methodology. For example, inspired by the CASP14 performance of AlphaFold2, last month a team of academics published their results showing their use of a ‘three-track neural network’ to obtain structure predictions with accuracies approaching those of DeepMind in CASP14.
Published just before the AlphaFold2 Nature papers came out, the approach – named RoseTTAFold – has performed well in predicting blind structures of 69 proteins and shows potential to predict complexes of unknown structure with more than three chains.
Final comments
Image: Protein crystals for X-ray crystallography.
It is important to recognise, as DeepMind scientists have done, the contribution of decades of work since the 1950s and 60s by structural biology researchers using wet lab methods (X-ray crystallography, nuclear magnetic resonance spectroscopy and now cryo-EM) to produce the body of high-quality, experimental protein structures and their deposition, curation and public availability via the Protein Data Bank (PDB) ‘as an enduring public good to promote basic and applied research and education across the sciences’.
Also to be highlighted I think is the role of the Structural Genomics Consortium (SGC) which has accelerated the output and open sharing – prior to formal publication – of protein structures and other reagents, ‘focusing explicitly on less well-studied areas of the human genome’.
These efforts have been critical enablers of AlphaFold’s subsequent success.
It’s important to emphasise that structural biologists will not be made redundant by AlphaFold2. They will be continue to be essential for fundamental and drug discovery research, including validating AI-predicted structures and pushing the boundaries of understanding how 3D structures enable the basic processes of life and disease.
And structural biologists will now incorporate AI methods to accelerate experimental protein structure determination, as in the case of the recent use of cryo-EM combined with deep learning-based structure prediction from AlphaFold2 to produce an atomic model for full-length SARS-CoV-2 Nsp2 protein – with implications for a functional role in RNA binding and potential application for drug design.
Overall, I believe that AlphaFold2 is a major advance along the technological journey of predicting the 3D structure of life’s proteins and that it will have a profound impact in accelerating our overall understanding of the fundamental structure-function basis of life and disease.
And it will contribute to a continued acceleration of the application of AI to drug discovery and biotechnology.
The journey continues.
Additional information
I found this podcast very interesting to listen to.
This video tells the AlphaFold story from DeepMind’s perspective.
Update - August 2022
In partnership with EMBL’s European Bioinformatics Institute (EMBL-EBI), AlphaFold announced just a few days ago (28 July 2022) that they are releasing the predicted structures for ‘nearly all catalogued proteins known to science’.
The release will dramatically expand the AlphaFold database by more than 200 times – from almost 1 million structures to more than 200 million structures.