-embed.jpg?sfvrsn=ab837b69_2)
Have you ever wondered how many different scientists it takes to develop a new drug?
From geneticists, biologists and chemists, to clinicians and industry scientists, drug discovery takes a lot of time and effort. It can involve working to identify the genes and proteins that are involved in a disease, finding out if they can be targeted by a drug, making new molecules that bind to your target, testing them to make sure they’re effective and safe… and all that’s before you even start clinical trials.
Drug discovery involves scientists from many different disciplines and fields, and it just wouldn’t be possible without chemists, biochemists, molecular biologists and many others working together.
I am a PhD student at The Institute of Cancer Research, London, working in both the Cancer Therapeutics Division on our Sutton campus, and the Structural Biology Division at our Chelsea site.
My project exemplifies the multidisciplinary nature of drug discovery, in that I’ve already found myself working in three different labs and learning lots of new techniques.
Investigating potential drug targets
My project involves trying to find new molecules that target interactions between proteins. Humans have thousands of proteins in our bodies, and each of these proteins has a different job to do to keep us alive and healthy.
Proteins can interact with each other, in vast signalling networks, and these signals tell cells in our bodies when to grow, or multiply, or even die when they are damaged.
In cancer, some of these processes become disrupted and unregulated, meaning cells keep growing and multiplying when they’re not supposed to.
A crystal structure of the protein Tankyrase. Molecules that bind to Tankyrase could be used to block the protein’s activity in cancer (image: Katie Pollock).
I’m looking for new molecules that bind to a protein called Tankyrase, to prevent abnormal cell signalling. Tankyrase can interact with many different binding partner proteins, and so is involved in a number of processes that become unregulated in different types of cancer.
For example, abnormal cell signalling is common in some types of bowel cancer. Mutations in Tankyrase binding partner proteins, or alterations in other proteins along the pathway, affect the signalling network, so studying how Tankyrase interacts with its binding partners could identify ways to stop these interactions, as a potential therapy.
Working at the interface of disciplines
Tankyrase is a member of a family of proteins called PARPs, and cancers with BRCA 1 and 2 mutations, such as some forms of ovarian cancer, can be treated with a PARP inhibitor called Olaparib. These drugs prevent signalling that the cancer cells have become reliant on to survive.
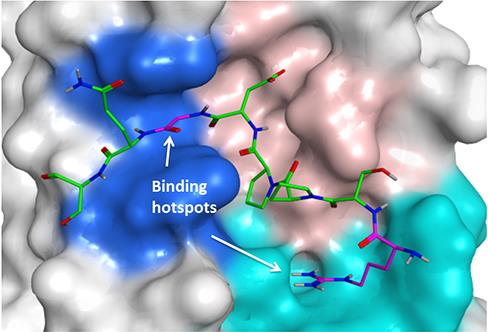
Tankyrase inhibitors are being developed that target an area of the protein called the catalytic domain, but I’m looking for molecules that target a region called the substrate binding domain, to investigate whether this approach could be an alternative method to inhibit Tankyrase function.
Trying to find inhibitors for the Tankyrase substrate-binding domain involves working at the interface of chemistry and biology. The chemistry side involves making peptides — strings of amino acids which form the building blocks of proteins — using structures based on the proteins that bind with Tankyrase.
Using an approach called peptide mimetics, we change one amino acid at a time to produce new compounds with properties more suitable for drug development.
On the biology side, I make proteins that resemble the Tankyrase substrate binding domain, and use a fluorescently tagged probe to see if my peptide mimetics can still bind to the protein.
My research is just a tiny part of the process of developing drugs, and already I’ve learnt so many new techniques in chemistry, biochemistry and biology, and as the project progresses it will become even more multidisciplinary.
By the end of my PhD I hope to have found some compounds that can be developed into tools to study Tankyrase binding, and maybe even a potential therapeutic treatment for cancer.
comments powered by