Background: Darwin’s branching tree of evolutionary phylogeny
08
Oct
2015
Since the turn of this century, cancer genomics has strongly endorsed the Darwinian view of cancer biology. Interrogation of the genomes of single cancer cells and multi-regional small biopsies of tumours have allowed us to construct evolutionary histories, or phylogenies, of cancer clones.
Posted on 08 October, 2015
by Mel Greaves

Charles Darwin’s Transmutation notebook B, 1837
Since the turn of this century, cancer genomics has strongly endorsed the Darwinian view of cancer biology 1,2. Interrogation of the genomes of single cancer cells and multi-regional small biopsies of tumours have allowed us to construct evolutionary histories, or phylogenies, of cancer clones – revealing genetic and cellular architectures very reminiscent of Darwin’s iconic drawing of evolution or speciation 2.
Scrutiny of cancer genomic sequences has also provided insight into the mutational processes involved – in some cases implicating particular exposures and revealing footprints of positive selection 3.
The result is that we now have a new perspective and understanding of cancer, one that reflects that fundamental law of biology: evolution by natural selection. The concept needs refinement and more detail – on clonal dynamics and ecosystem selective forces for example, but it’s becoming an integral part of mainstream cancer biology. It’s also highly relevant to the major challenges we face in understanding causation and effecting control or cure 4-6. We now appreciate that for a genetically diverse cancer population, drug treatment is likely to select for resistance as it does with pathogenic parasites, bacteria and viruses7. And, given the robustness and diversity that clonal evolution engenders, the sooner we intervene, to prevent cancer or forestall cancer evolution, the better.
Compare the figures below2,8-9 (click to enlarge) that appeared in recent important papers in this field, with the sketch in Darwin’s notebook above – quite remarkable!

- Yates LR and Campbell PJ, Evolution of the cancer genome. Nat Rev Genet, 2012, 13:795-806
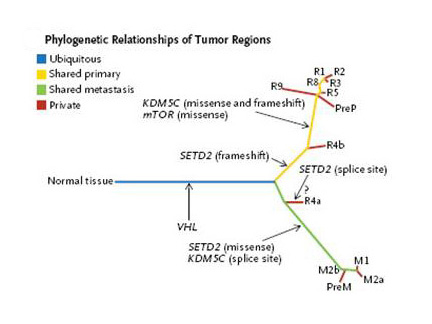
- Gerlinger M et al, Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. NEJM, 2012, 366:883-92

- Potter NE et al. (2013) Single-cell mutational profiling and clonal phylogeny in cancer. Genome Res, 23: 2115-2125. doi: 10.1101/gr.159913.113.
Mel
References
1. Yates LR, Campbell PJ. Evolution of the cancer genome. Nat Rev Genet 2012; 13: 795-806.
2. Greaves M, Maley CC. Clonal evolution in cancer. Nature 2012; 481: 306-313.
3. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature 2013; 500: 415-421.
4. Aparicio S, Caldas C. The implications of clonal genome evolution for cancer medicine. N Engl J Med 2013; 368: 842-851.
5. McGranahan N, Swanton C. Biological and Therapeutic Impact of Intratumor Heterogeneity in Cancer Evolution. Cancer Cell 2015 Jan 12; 27(1): 15-26.
6. Greaves M. An evolutionary foundation for cancer control. In: Stewart BW, Wild CP (eds). IARC World Cancer Report 2014. International Agency for Research on Cancer (IARC): Lyon, 2014, pp 337-345.
7. Goldberg DE, Siliciano RF, Jacobs Jr WR. Outwitting evolution: fighting drug-resistant TB, malaria, and HIV. Cell 2012; 148: 1271-1283.
8. Gerlinger M, Rowan AJ, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med, 2012; 366: 883-892.
9. Braggio E, Kay NE, et al. Longitudinal genome-wide analysis of patients with chronic lymphocytic leukemia reveals complex evolution of clonal architecture at disease progression and at the time of relapse. Leukemia, 2012; 26: 1698-1701.
comments powered by