
Charles Darwin's theory of evolution applies to all living things, including the process of how cancer cells can evolve resistance to drugs.
In the pursuit of more effective, tailored cancer therapies few challenges have proved more troublesome than treatment resistance — when cancer adapts to the drugs or radiotherapy we bombard it with and these interventions no longer work.
But now, scientists are increasingly excited about a new area of research that aims to slow or even halt the evolution of resistance. Among a number of potential leads, research to target a family of molecules called APOBEC proteins is causing a stir.
“We’re incredibly excited about this project,” said Professor Paul Workman, Chief Executive of The Institute of Cancer Research, London, who described the latest progress in this area at a session at the National Cancer Research Institute (NCRI) conference in Liverpool last week.
When we target tumours with drugs and radiation, many of the cancer cells die off. But some can develop adaptations that allow them to escape the deadly effects of our therapies. These cells survive and flourish at the expense of their neighbours, and the cancer is resurgent.
‘Survival of the nastiest’
This process of adaptation, evolution, and resistance — dubbed the ‘survival of the nastiest’ — is a major barrier to eliminating cancer and preventing relapse.
Overcoming evolution and resistance to treatment is an immense challenge, in part due to the sheer complexity of the intricate molecular networks that govern cells. Cancer cells can even ‘rewire’ their cellular processes to avoid succumbing to our treatments.
But the rewards, if we could find ways of overcoming evolution and resistance, could be huge. Which is why there is such interest in the latest research to tackle the problem.
Overcoming cancer evolution is at the heart of the ICR’s and The Royal Marsden’s new research strategy.
Projects involving APOBEC proteins are at the forefront of this new approach to cancer research and treatment.
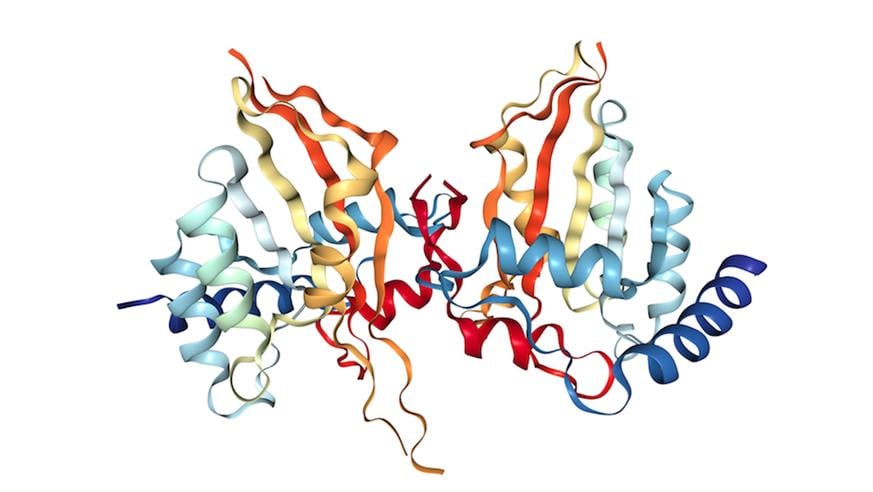
Crystal structure of human APOBEC3A protein. Structural information obtained from RSCB Protein Data Bank and visualized with the NGL protein viewer.
APOBEC proteins have roles in our immune system battling viruses, but when these go wrong they can mutate our own DNA, leading to cancer. The mutational ‘footprint’ left by a subset of this family, APOBEC3, has been detected in half of cancer classes. In breast cancers, tumours with high levels of APOBEC3B were found to carry twice as many mutations as those with lower levels. APOBEC-driven mutations are also more common in ‘older’ cancers, where they drive further tumour evolution and increase the likelihood of resistance.
So scientists are aiming to find ways of blocking the ability of APOBEC3B to cause damage to DNA, in order to slow the pace of tumour mutation and so delay resistance to therapy. That’s the theory, anyway.
ICR researchers have made significant progress investigating the structure of the protein using a range of imaging approaches. The hope is this research can lead to drugs that can enter clinical trials in the not-too-distant future.
Many targets remain
And there may be many other potential targets that could be ‘drugged’ to achieve the aim of arresting the growth of resistance.
Of the 500 or so cancer genes now identified and validated, Professor Workman explained, we have developed drugs for only 5%. “Ninety-five percent of cancer gene targets…remain to have any kind of chemical inhibitors working on them,” he said.
So there is great potential for drugs acting on innovative new targets. One such promising group — called the Hsp90 inhibitors — are also attracting great interest.
Hsp90 is a type of molecular chaperone — a protein that helps other molecules carry out cellular processes. Blocking its action can hit multiple vulnerabilities in cancer cells. Research suggests this may have the potential to overcome and prevent drug resistance, for instance to drugs called kinase inhibitors or hormonal treatments.
The ICR discovered one such Hsp90 inhibitor, called luminespib, with its partner Vernalis and licensed it to Novartis to take it from the lab to the clinic. There are now promising results in clinical trials of the drug against tumours such as breast and lung cancers, and especially one particular set of lung tumours with a genetic driver called 'EGFR exon 20 insertion'.
Boosting survival rates
Professor Workman hopes this and other novel therapeutics targeting adaptive mechanisms and cancer evolution can “improve overall survival and cure rates as we so desperately need”.
Looking to the future, a big project at the ICR is to develop what is called ‘dynamic adaptive therapy’. One day, Professor Workman explained, a combination of therapies — potentially including Hsp90 and APOBEC inhibitors — could be administered early in a treatment plan to delay resistance, giving clinicians a better chance of successfully treating a cancer. And therapy would be adapted in response to how a patient’s cancer responded throughout their treatment.
Driving this will be the explosion of ‘Big Data’ approaches, artificial intelligence and machine learning — all areas the ICR is investing in as part of its new Department of Data Science. Plugging in patient data - from genomics to clinical data, imaging and even information from the patient’s GP — will generate recommended treatment combinations tailored to the individual, with the ultimate goal of better outcomes for the patient. Plans are taking shape to start trials of novel dynamic adaptive and individualised therapies over the next few years.
Our scientists at the ICR and collaborators are working hard to ensure this becomes a reality. Overcoming the major challenge of evolution and resistance would be a giant step forward in cancer care.
Other posts from NCRI 2016
comments powered by