My colleagues and I have just published what we think is a useful case study in attempting to find small molecules that target a biologically important but extremely hard-to-drug protein.
This is a story of several years of research that generated useful information on the target protein and also identified very early, mechanistically interesting prototype inhibitors – but where we nevertheless concluded that any future progression to more potent chemical probes and drugs would be extremely challenging.
In my previous post I discussed how in small-molecule drug discovery it’s essential to consider not only the importance and biological validation of a given drug target in the disease of interest, but also the extent to which it is technically druggable.
I highlighted how it’s so important to make a critical assessment of the protein target – especially an analysis of its 3D protein structure. And I emphasised how a healthy dose of realism is needed about the likely scale of the challenge and that technically tougher targets will require perseverance and ingenuity – recognising also that many projects are not successful in coming up with a drug.
In addition, I stressed how drug discovery builds on past successes and learns from earlier failures. So it’s essential that researchers publish and share widely their experiences in tackling the hard-to-drug targets – including the unsuccessful as well as the successful stories.
For that reason, I discuss here our recent experience of a project where our team of researchers worked for four years, made significant progress and gained new knowledge – but in the end decided to call a halt and share our experience to help others.
First a health warning to readers: this post is of necessity more technically detailed than the previous one. I think it’s likely to be of most interest to practising scientists or people involved in other ways in the challenge of drug discovery and development. This includes biologists interested in proposing a potential new target.
Different ways to drug Hsp90
The potential drug target that we chose to investigate was the interaction between a protein called Hop and the molecular chaperone Hsp90.
Hsp90 is a type of protein called a 'molecular chaperone' that cells rely on very heavily for ensuring that certain other proteins – referred to as ‘client’ proteins – remain correctly folded, active and stable in cells. In terms of its biological attractiveness as a drug target, Hsp90 plays a role in maintaining various client proteins that have an important role in disease.
In particular, it was recognised that blocking Hsp90 effectively could offer an exciting new way to treat cancer, through depletion of cancer-causing client proteins, and also potentially other diseases where Hsp90 plays a role – notably Alzheimer’s disease and viral infections, including COVID-19. Based on this concept, there have been extensive efforts in academia and the biopharmaceutical industry to discover small-molecule Hsp90 inhibitors.
In earlier work – and following evidence obtained with natural products – we and others were successful in discovering synthetic small-molecule inhibitors that bind to the very tractable ATP pocket of Hsp90 located in the N-terminal domain. These inhibitors – including our own ICR/Vernalis drug luminespib, also known as AUY922 – have shown clinical activity, especially in breast and lung cancer, although none has yet been approved.
Limitations of the N-terminal ATP pocket Hsp90 inhibitors include side effects and also the induction of the heat shock response mediated by Heat Shock Factor 1 (HSF1), which blunts their anticancer effect.
Since the activity of Hsp90 is controlled by many interacting proteins, known as ‘co-chaperones’, a number of researchers, including ourselves, have been interested in inhibiting Hsp90 co-chaperones as an alternative therapeutic approach. For example, we have discovered inhibitors of Hsp70 family proteins, which collaborate with Hsp90. And we have also made good progress in discovering inhibitors of the HSF1 pathway.
Targeting the interaction between Hsp90 and Hop
In our newly reported research, we set out to target the protein-protein interaction between Hsp90 and the co-chaperone Hop – which is one of the many Hsp90 co-chaperones containing the ‘tetratricopeptide repeat’ or TPR structural motif.
This motif comprises 34 amino acids and has been widely used in the evolution of more than 5,000 proteins to provide scaffold or adaptor functions to mediate protein-protein interactions and thereby achieve a wide range of beneficial biological effects in cells.
Finding chemical probes and drugs for TPR domains of proteins is therefore of more general interest beyond the specific case of the Hop-Hsp90 protein-protein interaction.
A recent 3D structure of the complex formed between Hop and Hsp90 – obtained by cryo-EM – shows that it stabilises an open state of Hsp90 in which it is poised for client protein loading by Hsp70 and the subsequent N-terminal dimerisation and ATP hydrolysis that is required to complete the chaperone cycle.
Since Hop regulates the chaperone function of Hsp90, we reasoned that the protein-protein interaction that the TPR2A domain of Hop forms with the MEEVD peptide at the C-terminal tail of Hsp90 would be an interesting alternative site for potential inhibitors.
Furthermore, we believed that inhibitors acting on the Hop TPR2A domain could result in interesting biological effects and potential therapeutic mechanisms distinct from ATP site binders like luminespib. One such beneficial effect could be the inhibition of HSF1 activity.
Our study was carried out as a Wellcome-funded PhD project by Dr John Darby – a very talented then-graduate student who is now at the University of Sheffield – supervised by myself and my long-term structural biology colleague Professor Laurence Pearl, working both at the ICR and the University of Sussex. Several collaborators at the ICR and Imperial College London also made invaluable contributions.
This type of multidisciplinary collaboration between the ICR and Imperial is now being intensified in our joint CRUK Convergence Science Centre.
Severe druggability challenge called for multiple technical approaches
Recognising that finding inhibitors of the TPR2A-Hsp90 interaction was probably going to be very difficult, we called on the help of Professor Bissan Al-Lazikani to carry out a very detailed druggability analysis – using the sophisticated methodology, incorporating machine learning and AI prediction, that is available in our canSAR BLACK knowledgebase.
canSAR contains assessments of more than 118,000 structures of complexes containing more than 646,000 protein-interfaces, and approximately 80,000 of these cavities are rated as highly druggable. In contrast, we found that none of the 13 available Hsp90/Hsp70-TPR domain structures from eight distinct complexes, including the Hop TPR2A domain, was predicted to be druggable using multiple standard small-molecule criteria.
The protein-protein interaction-mediating surface groove in the TPR2A domain scores poorly on a variety of druggability criteria. Crucially, the clusters of positively charged groups that are located in the shallow channel on the Hop TPR2A domain which interacts with the Hsp90 C-terminal MEEVD peptide present a special challenge for druggability, since the acid groups that would be needed for the binding of a small molecule would be likely to be a limitation for membrane permeability.
Recognising the scale of the challenge, we undertook a number of orthogonal approaches – the idea being that different technical methods would give us more shots on goal and thus mitigate the risk of individual ones failing to deliver.
With assay design help from Swee Sharp in our lab, John Darby used an Alpha screen binding method – which measured the interaction between the recombinant TPR2A domain and the Hsp90 MEEVD peptide – to run a high-throughput screen of an ICR library of around 80,000 diverse ‘lead-like’ compounds. This compound collection has previously generated progressible hits against multiple protein targets at the ICR.
However, in this case we did not identify any advanceable hits – only false positives that we eliminated in follow-up assays.
We also screened 2,000 low molecular weight ‘fragment-like’ compounds. An advantage of this approach is that since the fragments are very small – typically less than 200 Da in molecular weight – they are less likely to contain groups that prevent binding and more likely to identify functional motifs that successfully match up to the structural requirements of the target. But again we found no hits – certainly a further warning sign about the tractability of the target.
Solving the NMR solution structure of the Hop TPR2A domain
Recognising the importance of knowing the 3D protein structure in the design of ligands, probes and drugs, an important feature of our publication is that we report a new apo, solution-state nuclear magnetic resonance (NMR) structure of recombinant Hop TPR2A.
This was solved in collaboration with Dr Peter Simpson and Professor Steve Matthews at Imperial College London. The structure is shown in the figure below.
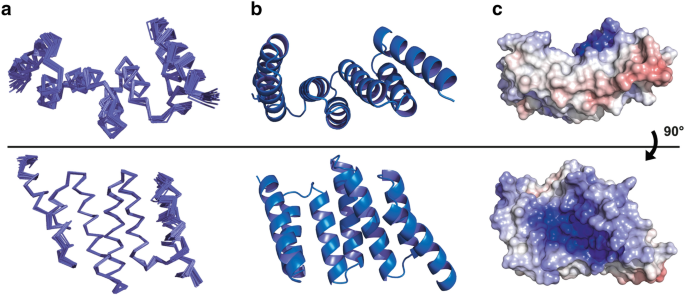
Caption: NMR structures of the Hop TPR2A domain. a) Ensemble views of the final 20 models; b) Ribbon representation of the ensemble average; c) Electrostatic potential of the solvent-accessible surface – the dark blue colour illustrates the shallow, strongly positively charged groove that is a particular challenge for druggability. Reproduced from Darby et al; PDB:2NC9.
A novel feature from a structure/function point of view is that we found the Hop TPR2A domain to be fully folded in solution, even in the absence of the Hsp90 C-terminus.
In addition to the basic research interest, our NMR solution structure would prove to be very valuable in the later phase of the search for inhibitors.
Discovering carboxylate clamp mimetic HopTPR2A binders
Given the lack of hits from the high-throughput screening of lead-like and fragment libraries – and faced with a very substantial druggability challenge – we next took inspiration from the native ‘carboxylate clamp’ interactions known from the X-ray structure to be crucial in the binding of the MEEVD peptide of Hsp90 to Hop TPR2A domain.
The carboxylate clamp involves two carboxylic acid residues present in Hsp90 MEEVD peptide that anchor into the positively charged channel of the Hop TPR2A domain.
We therefore adopted a ‘ligand-directed approach’, seeking to find acidic compounds that would act as mimics to compete with the Hsp90 MEEVD peptide for binding to the positively charged TPR2A groove.
Working in partnership with Dr Lewis Vidler – now at UCB, Slough, and then a Cancer Research UK-funded PhD student in the lab of Professor Swen Hoelder at the ICR – John Darby selected simple compounds containing the succinic acid substructure that contains two carboxylic acid residues. He then tested these for their ability to block the interaction between Hsp90 MEEVD peptide and the Hop TPR2A domain using a LANCE method.
We were very pleased to obtain positive results which provided encouraging proof of concept that simple diacid-containing fragments could mimic the carboxylate clamp mechanism and disrupt Hsp90 MEEVD peptide binding to the Hop TPR2A domain – albeit requiring high, millimolar concentrations of the compounds at this stage.
Building on this proof of concept, Lewis then carried out a focused in silico screen of more than 4 million commercially available compounds. Our aim was to identity compounds that could maintain the carboxylate clamp mechanism of binding while also making additional hydrophobic (‘water-hating’, non-polar) contacts with the TPR2A domain – as also used by the native Hsp90 MEEVD peptide – and thus achieve greater potency.
The initial output of the in silico screen yielded 155 hit compounds. Lewis and John inspected how these hits docked into the Hsp90 MEEVD-binding channel of Hop TPR2A and selected 14 for biochemical testing. We were again very pleased that one of the hits (referred to as compound 7) showed about 10-fold more potent activity than the simpler diacids – exhibiting 50% inhibition at 74 micromolar (known as the IC50 value).
We also studied some simple structural analogues of compound 7 – which is based on the tryptoline scaffold and has two carboxylic acid residues – and defined the structure-activity relationships. Interestingly, analogue 8 has only one carboxylic acid residue but still retains activity, with a potency only four times lower than compound 7.
Our molecular docking studies suggested that compound 7 may bind to one side of the MEEVD peptide-binding channel on Hop TPR2A domain and also make the additional, desired hydrophobic contacts. This is illustrated in the figure below.
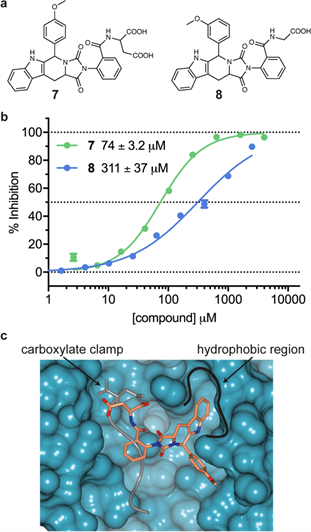
Caption: a) Chemical structure of compound 7 from the carboxylate clamp-inspired, ligand-directed in silico screening studies and the close analogue compound 8. Note that 7 is based on the tryptoline scaffold and has two carboxylic acid residues whereas analogue 8 has only one; b) Concentration-response curves for inhibition of the Hsp90 MEEVD peptide binding to the Hop TPR2A domain with IC50 values shown for 7 and 8; c) In silico docking pose for compound 7 (in coral) against the Hop TPR2A (turquoise surface) with the Hsp90 MEEVD peptide (in grey) overlaid for comparison. The carboxylate clamp and hydrophobic binding region are labelled. Reproduced from Darby et al.
The molecular structure shown above is a computer model and so we wanted to check experimentally that our inhibitor compounds were indeed binding in the way the model suggested. This is where our NMR solution structure of the Hop TPR2A domain comes into the story once again.
We were able to use our NMR structure to detect binding of our inhibitors to the Hop TPR2A domain by measuring what are known as the protein-detected NMR chemical shift perturbations (CSPs). These NMR CSPs reflect the effects of the Hsp90 MEEVD peptide or small-molecule inhibitor on the immediate chemical environment of the binding site – and hence reveal the actual binding mode.
We were reassured and delighted when the results did indeed confirm our proposed mode of interaction for succinic acid and tryptoline compounds with the binding channel on Hop TPR2A. This indicated specific binding in the groove occupied by the MEEVD peptide, including the carboxylate clamp region.
Conclusions, learning and future prospects for hard-to-drug targets
Overall, our work provides: 1) a novel NMR structure of the Hop TPR2A domain in solution; 2) a detailed and rigorous assessment of druggability showing that the Hop TPR2A domain is a major challenge; 3) robust orthogonal screening approaches; and 4) prototype inhibitors that mimic the native carboxylate clamp interaction required for the binding of C-terminal Hsp90 MEEVD peptide to the Hop TPR2A domain.
We highlight in our publication, that our work could offer a potential way forward to address the highly challenging nature of the Hop TPR2A target – with relevance to other TPR domain interactors as well.
On the other hand, even our best ligands exhibited only moderate potency. They also contain an acid, which can create some problems for cell permeability, even though a number of drugs with acidic residues have been approved.
We are very honest about the scale of the challenge. We point out that progress would require a major effort to identify more potent tool compounds and potential drugs. Although such effort could potentially be justified given the biological significance of Hsp90-TPR interactions, we nevertheless caution that such a campaign would be very difficult.
And at the end of the Discussion section of our publication, we state: ‘We recognise that the limited druggability of TPR domains may ultimately preclude the development of high-affinity ligands.’
Not only was this project a four-year, full-time journey for John’s PhD, but it also had the input of many colleagues and collaborators. We felt it was very important to share our experience of working at the edge of current technology.
Some journals we originally submitted the paper to, while recognising that the work was rigorous and technically sound, did not give it a high priority because the druggability challenge suggested it was unlikely that the inhibitors could be progressed further.
This is a common problem: publication bias in favour of those projects that succeed in progressing to probes and drugs. This bias gives an unrealistic picture of the likelihood of success in drug discovery, leading many biologists to have unrealistic expectations for their target of interest.
We are pleased that our experience has now been published in Scientific Reports so that the learning points are widely and freely available. Publication of our results will allow other researchers either to walk away from TPR domain targets, or to take them on in full knowledge of the scale of the challenge.
There are now many promising new approaches to drugging challenging targets. At the moment, for instance, there is great enthusiasm for targeted protein degradation – a recent example being the ICR spin-out company Monte Rosa.
The advantage of this exciting approach is that even relatively weak ligands can be harnessed to drive the depletion of the target protein in the cell, and the binding pocket does not need to have functional significance for the target protein.
Another current approach is to use targeted inhibitors of transcription to switch off the production of ‘undruggable’ or very hard-to-drug target proteins. Led by ICR's Professor Louis Chesler and also by Professor Charles Lin – the latter being then at Baylor College of Medicine and now at Kronos Bio – we have just published preclinical results showing very promising therapeutic activity of the CDK9/2 inhibitor fadraciclib in mouse models of the aggressive childhood cancer neuroblastoma driven by the NMYC transcription factor – a disordered protein that has proved impossible to drug directly so far.
Fadraclib, which was discovered in a collaboration between the ICR and Cyclacel, is effective in this setting as a result of the inhibition of CDK9 decreasing the expression of the NMYC gene, leading to apoptosis through the simultaneous combined effect of CDK2 blockade, together with targeting of the adrenergic neuroblastoma cell state.
These studies have underpinned the upcoming clinical trial of fadraciclib in paediatric NMYC-driven neuroblastoma. I plan to return to this story in a future blog.
Standing back from the Hsp90 study and all in the wider context, I think the general message is that with perseverance and ingenuity progress can be made on very difficult targets, but there is no guarantee of success.
Researchers need to take a long hard look at hard-to-drug targets and to assess the benefits and risks, and the effort that would be needed to succeed.
Having a 3D protein structure is a big help. Slowly but surely, some of the apparently 'undruggable' targets are now being cracked.
Sharing information on the unsuccessful as well as the successful experiences in drugging such important but technically challenging targets is essential to help us move the whole field forward.
comments powered by