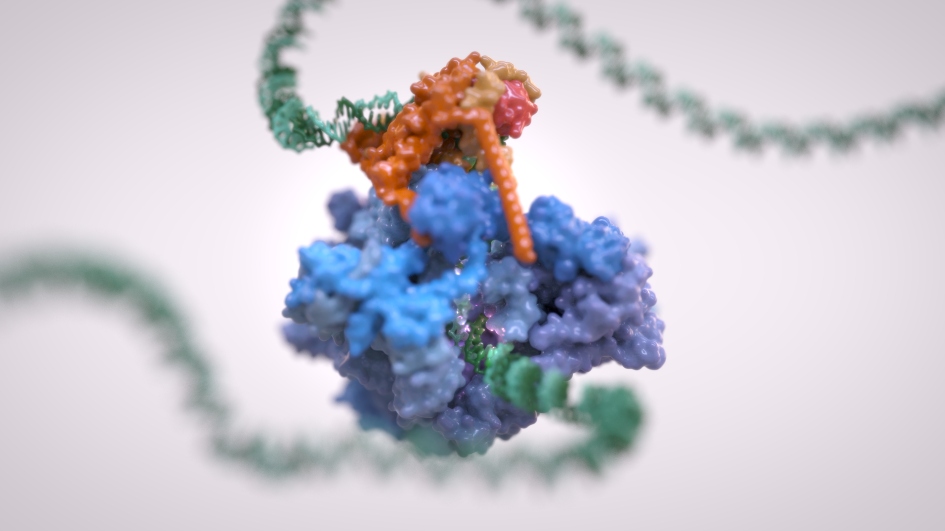
Image: RNA Polymerase III. Credit: Alessandro Vannini/Jeroen Claus, Phospho Biomedical Animation
Scientists have created a three-dimensional map of a complex of molecules that plays a fundamental role in life – and which when it goes wrong is linked to increased sensitivity to viral infections and neurodegenerative diseases, and potentially also to cancer.
Their research has revealed the structure of the protein complex in human cells in stunning detail and shown precisely how this is affected by different mutations. It is set to act like a treasure map for guiding future research, and could lead to new targeted drugs for a range of diseases. In addition to neurodegenerative conditions and viral infection, the researchers also believe that there is a link between altered regulation of the protein complex and cancer.
The protein complex, known as RNA polymerase III or Pol III, reads DNA to decode ‘housekeeping’ genes and helps create the proteins that form the basic building blocks of cells. Cancer cells often hijack this process to fuel their rapid growth and division.
Capturing the detailed big picture of the protein complex
Three years ago, scientists at The Institute of Cancer Research, London, unveiled the structure of Pol III in yeast – describing it as akin to a Van Gogh painting due to capturing the detailed big picture of the protein complex.
Now the same team of scientists, working with colleagues in Germany, has revealed the three-dimensional structure of the human version of the protein complex – as well as mapping the precise structural effects of a wide range of disease mutations relating to neurodegeneration and increased sensitivity to infection by viruses. This could guide the way towards efforts to target the protein complex with new drugs.
The study, funded by Wellcome and Cancer Research UK, is published in Nature Communications today.
The team at The Institute of Cancer Research (ICR) made use of two Nobel Prize-winning scientific techniques – Cryo-EM, which won the Nobel Prize in Chemistry in 2017, and gene editing with ‘CRISPR’ – which won the Nobel Prize in Chemistry in October this year.
Precise, high resolution structure
Applying these two techniques to human cells in the lab, the researchers created many detailed images of the protein complex in its healthy form, and when affected by various mutations. They then grouped these together to obtain a precise, high-resolution structure of the protein machinery in action or when malfunctioning.
Researchers believe cancer cells take advantage of Pol III to keep them fuelled with the protein building blocks they need for rapid growth and division, while mutations in Pol III can also lead to neurodegenerative disease in humans. Researchers can now explore in the lab whether this is a promising therapeutic target in cancer.
By being able to visualise the 3D structure of human Pol III in detail, the researchers managed to map more than 85 per cent of known genetic mutations associated with Pol III very precisely. This allowed researchers to narrowly define the areas where these mutations occur and helped them interpret the mutations’ effects at a molecular level.
They found that mutations affecting the development of the central nervous system, for example, tend to cluster in specific hotspots, often between the same subunits of the protein complex.
Could be used to help design new drugs
The findings offer information on the potential effect that mutations could have on the binding of potential drugs. Drugs work by binding to protein targets, so the new structural information could eventually be used as a 3D map to help design new drugs.
The study confirms the overall similarity between the human version of Pol III and its yeast counterpart, but also highlights specific structural differences that weren’t known before and are of functional significance.
Study leader Professor Alessandro Vannini, Team Leader and Professor of Integrative Structural Biology at The Institute of Cancer Research, London, said:
“The Pol III protein complex is fundamental to life – decoding our DNA and ensuring our cells can build the proteins they need to grow and divide. And we believe it could have a role in the development of cancer.
“Our findings provide a vivid, extremely detailed three-dimensional map of Pol III, showing the structure of its various active sites, and how these are affected by various mutations linked to neurodegenerative diseases and viral infections. We hope our work can act as a 3D treasure map for the future discovery of new treatments targeting the Pol III complex.”
First author Dr Ewan Ramsay, Post Doctoral Training Fellow at The Institute of Cancer Research, London, said:
“We used very exciting techniques in this study – a revolutionary type of microscopy called Cryo-EM and a technology to edit genes known as CRISPR. This allowed us to create and visualise the human structure of the extremely important protein complex Pol III, which hadn’t been done before.
“Both of these techniques have won Nobel Prizes in the last three years and continue to revolutionise the field of structural and cell biology.”
Professor Paul Workman, Chief Executive of The Institute of Cancer Research, London, said:
“This exciting study reveals how a vital component of life functions in human cells – and crucially how it malfunctions too.
“This is a fantastic example of the importance of fundamental or ‘basic’ science. Understanding the inner workings of cells is an essential foundation for determining what goes wrong in cancer, neurodegeneration and other diseases, paving the way for potential new treatments.”